
Biofunctional study on chemoresistance in esophageal squamous carcinoma cells induced by missense mutation of NOTCH1 p.E450K
Highlight box
Key findings
• The NOTCH1 p.E450K point mutation causes chemotherapy resistance in KYSE140 and KYSE450 esophageal squamous cell carcinoma (ESCC) cells.
What is known and what is new?
• The detection rate of NOTCH1 missense mutations was significantly increased in patients with ineffective neoadjuvant chemotherapy (nCT). Further study showed that NOTCH1 p.E450K was the only mutation that was being repeatedly detected.
• We elucidate the biological and functional changes in cell lines with a NOTCH1 missense mutation and systematically analyze the effect of the NOTCH1 p.E450K missense mutation on the drug resistance of ESCC cells.
What is the implication, and what should change now?
• ESCC is a malignant tumor with high incidence and strong invasiveness. nCT combined with surgery is one of the main strategies for the treatment of resectable locally advanced ESCC. Although nCT has achieved good effects, resistance of tumor cells to chemotherapy is still a problem. If the mechanism underlying the involvement of the NOTCH signaling pathway in cell drug resistance is elucidated, it may provide new therapeutic targets and targeted pathways for molecular targeted therapy for the reversal of drug resistance in esophageal cancer. This missense mutation may be a potential predictor of clinical chemotherapy efficacy and an important target for the reversal of drug resistance, which is worthy of further study.
Introduction
Esophageal cancer is a common malignant tumor of the gastrointestinal tract. In 2020, the incidence and mortality of esophageal cancer worldwide ranked eighth and sixth, respectively, among all malignant tumors (1). The main pathological types of esophageal cancer are esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). More than half of ESCC cases worldwide are in China (2).
Neoadjuvant chemotherapy (nCT) combined with surgery is one of the main strategies for the treatment of resectable locally advanced ESCC. However, nearly 40% of patients do not benefit from nCT, and approximately 5% even experience progression (3), suggesting that these patients may have drug resistance mutations. In response to the chemotherapy resistance of some tumors, many scholars have conducted screening studies on drug-resistant mutations in the hope of improving the efficacy of chemotherapy (4). Some studies have shown that the abnormal activation of the NOTCH signaling pathway is closely associated with the occurrence, development, invasion, and metastasis of many tumors (5,6). It has been confirmed in esophageal cancer and other tumors that the activation of the NOTCH1/HES1 pathway can lead to tumor chemotherapy resistance (7,8). However, most of these studies addressed the regulation of NOTCH1 transcription or expression and the subsequent abnormal activation of the NOTCH1/HES1 pathway leading to drug resistance. To date, no studies have shown that NOTCH1 missense mutations affect the ligand binding status of receptors in this pathway to cause drug resistance. A previous study (9) on resistance to nCT in esophageal cancer showed that the detection rate of NOTCH1 missense mutations was significantly increased in patients with ineffective nCT. Further study showed that NOTCH1 p.E450K was the only mutation that was being repeatedly detected. Our previous study showed that the NOTCH1 p.E450K (p.Glu450Lys) missense mutation may lead to tighter binding of the mutant protein to the DLL4 receptor, thereby continuously activating downstream signaling pathways (10). Missense mutations may alter drug effects by affecting cell signaling pathways. Based on this, we used a series of experiments to investigate the effect of a NOTCH1 missense mutation on cell phenotype, to elucidate the biological and functional changes in cell lines with a NOTCH1 missense mutation and to systematically analyze the effect of the NOTCH1 p.E450K missense mutation on the drug resistance of ESCC cells. We present this article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1880/rc).
Methods
Cell lines and Sanger sequencing
ESCC cell lines KYSE140, KYSE150, KYSE410, KYSE450, and KYSE510 (from the Hormel Research Institute of Henan Cancer Hospital, a donation from Professor Kangdong Liu) were chosen as candidate cells. The DNA of relevant cells was extracted and subjected to polymerase chain reaction (PCR) amplification and electrophoresis, and after purification, sequencing screening was performed. According to the sequencing results, KYSE140 and KYSE450 cells were selected as the research objects of this experiment. Sanger sequencing was performed at BGI Genomics (Shenzhen, China).
Construct of point mutation cell lines
The KYSE140 cell line and KYSE450 cell line were chosen as the research objects of this study, and CRISPR/Cas9 technology was used to establish the KYSE140 and KYSE450 point mutation cell lines. This experiment was partly completed by Beijing Biocytogen Biotechnology Co., Ltd. (Beijing, China).
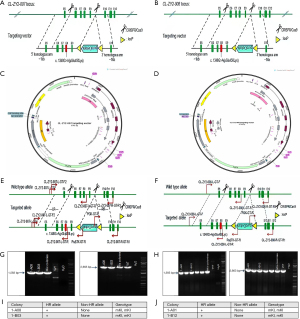
Construction of a point mutation cell line was first conducted using the KYSE140 cell line (Figure 1). The point mutation c.1348G>A (pGlu450Lys) was knocked-in to exon 8 of the NOTCH1 gene. After target sequencing was confirmed, the single-guide RNA (sgRNA) recognition sequence was completely consistent with the DNA sequence of the KYSE140 cell line. According to the design principles of sgRNA, at the 5' target locus, eight sgRNAs were designed for the 3' target site region. After validation, sgRNA5 and sgRNA12 were selected and electrotransformed into the KYSE140 cell line with the targeting vector. After drug screening, positive clone enrichment, and PCR screening, two positive clones were obtained. Then, PCR product sequencing confirmed that the KYSE140 cell line was homozygous for the knock-in point mutation. The KYSE450 point mutation cell line was prepared using the same approach as that used for the KYSE140 cell line, and two positive clones were obtained.
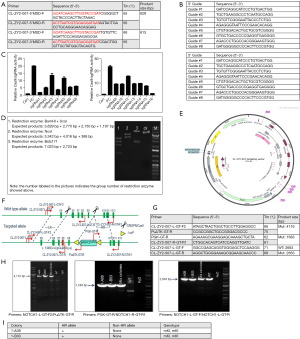
Drug sensitive test
The above groups of cells in the logarithmic growth phase were digested by trypsin (Servicebio G4004, Wuhan, China), seeded in 96-well plates at 5×103 cells per well, and incubated for 12 h. Then, cell suspensions were treated with cisplatin (0, 3.1, 6.3, 13, 25, 50 µg/mL) or paclitaxel (0.8, 1.6, 3.1, 6.2, 12.5, 25, 50, 100, 200 µg/mL). After 0, 24, and 48 h of treatment, 10 µL of Cell Counting Kit-8 (CCK-8) reagent (Vazyme, Nanjing, China) was added to each well. After the addition of the CCK-8 reagent, the cells were incubated for another 4 h. Optical density (OD) was measured with a microplate reader (Perlong, Beijing, China). Cell survival rate was calculated as (OD value of experimental group − OD value of blank group)/(OD value of control group − OD value of blank group) × 100%. The half-maximal inhibitory concentration (IC50) was calculated using GraphPad Prism 5.0 software to study and evaluate the sensitivity and resistance of cells to cisplatin/paclitaxel.
Wound scratch assay
A marker pen and ruler were irradiated with UV light for 30 min on a clean bench. A horizontal line was drawn on the back of each six-well plate, and approximately 1×105 cells were added to reach confluence overnight. The cell layer was scratched on the second day. After scratching, the cell layer was washed with phosphate-buffered saline (PBS) and incubated in fresh serum-free culture medium in an incubator (37 ℃, 5% CO2). Then, the cells were removed from the incubator at 0 and 24 h and observed under a microscope (quadruple field of view). The scratches were photographed, and the area of the scratch was calculated (UOP DSY5000X, USA). After opening the images in ImageJ, eight horizontal lines were randomly drawn, and the mean value of intercellular distances was calculated.
Transwell cell migration and invasion assay
Cells were digested with trypsin, counted, and adjusted to an appropriate density. In a 24-well Transwell plate (NEST 0223A, China), 400 µL of complete medium was added to the lower chamber, and 200 µL of serum-free cell suspension (NEST 725301, China) was added to the upper chamber. After culturing for 24 h in an incubator, the cells were fixed in paraformaldehyde for 25 min, washed three times with 0.9% NaCl, and stained with crystal violet for 40 min. A multifunctional camera was used to take pictures, and differences between the groups were observed. The measurement values were recorded, and the results were analyzed using GraphPad Prism 5.0 software.
Cell proliferation assay
Cell proliferation was evaluated by the CCK-8 assay. A total of 1×103 KYSE140-wild-type (WT)/mutant-type (MT) or KYSE450-WT/MT cells were seeded in each well of a 96-well plate. After overnight incubation, absorbance value was read at a reference of 450 nm. The inhibition rate (%) was calculated as (1 − OD value of treated group/OD value of the control group) × 100%.
Flow cytometry
Propidium iodide staining was used to assess the cell cycle status of the ESCC cell lines in the absence and presence of the NOTCH1 p.E450K point mutation. Cells in each group were digested with trypsin, centrifuged and fixed in 75% ethanol, then, propidium iodide staining solution was added and the cells were incubated at room temperature for 30 min in the dark, stored at 4 ℃ in the dark and analyzed within 24 h. A flow cytometer (BD Biosciences Canto II, San Jose, CA, USA) was used to detect red fluorescence at an excitation wavelength of 535 nm and an emission wavelength of 615 nm and light scatter. Cellular DNA content analysis and light scatter analysis were performed using FlowJo analysis software (FlowJo10.2).
The apoptosis rates of the ESCC cell lines in the absence and presence of the NOTCH1 p.E450K point mutation were determined using an Annexin V-fluorescein isothiocyanate (V-FITC)/propidium iodide (PI) kit (Beyotime C1062S, Shanghai, China). Briefly, the cells were digested, washed, and centrifuged in cold PBS. Staining was performed with Annexin V-FITC and PI at room temperature (15 min). Flow cytometry (BD Biosciences Cantoll) was performed to analyze cell apoptosis. Data were quantified using FlowJo analysis software (FlowJo10.2).
Statistical analysis
The data were statistically analyzed and compared using the statistical software SPSS 23.0 (IBM, Armonk, NY, USA). Each experiment was repeated at least three times, and P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was reviewed and approved by the Ethics Review Committee of the Affiliated Cancer Hospital of Zhengzhou University/Henan Cancer Hospital (No. 2021-KY-0049-001).
Results
Sanger sequencing screening
Sequencing results showed that the NOTCH1 p.E450K point mutation was located in NOTCH1-E8 and that KYSE140 and KYSE450 cells did not have exon mutations in the NOTCH1 ligand binding region in E8. KYSE140 and KYSE450 cells were chosen as the research objects of this study.
Construction of point MT cell lines (CRISPR/Cas9 technology)
The NOTCH1 gene is approximately 51.4 kb in length and is located on the anti-strand on chromosome 9 (NCBI ID: 4851). After drug screening, the enrichment of positive clones, and PCR screening, two positive clones were finally obtained. PCR product sequencing showed that 1-A08, 1-B03, 1-A01, and 1-B12 were homozygous knock-in clones of the KYSE140/450 cell lines. The construction and validation of the NOTCH1 c.1348G>A (pGlu450Lys) point mutation in the two cell lines are shown in Figure 2.
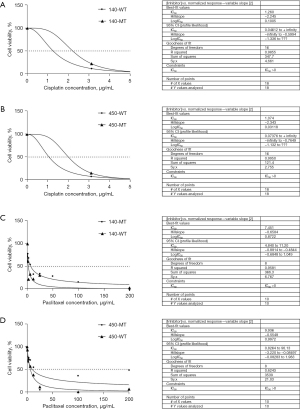
NOTCH1 p.E450K point mutation promoted chemotherapy resistance
To study the effect of the NOTCH1 p.E450K point mutation on chemotherapy resistance in ESCC cell lines, the KYSE140/450 cell lines were treated with cisplatin or paclitaxel.
The inhibition results for the KYSE140-WT/MT and KYSE450-WT/MT cell lines after treatment with different concentrations of cisplatin showed that with increasing concentrations of cisplatin, the inhibitory effect on the KYSE140-WT/MT cell line and the KYSE450-WT/MT cell line gradually increased in a dose-dependent manner, the IC50 value of the MT cell line KYSE140-MT/KYSE450-MT was higher than that of the WT cell line KYSE140-WT/KYSE450-WT. Under the same cisplatin concentration, the inhibitory effect of cisplatin on the MT cell line was always weaker than that on the WT cell line (Figure 3A,3B), and the difference was significant (P<0.001). This suggests that the NOTCH1 p.E450K point mutation can reduce the sensitivity of ESCC cell lines to chemotherapeutic agents, which in turn leads to chemoresistance, in terms of IC50, the same conclusion was obtained in the paclitaxel-treated group (Figure 3C,3D).
NOTCH1 p.E450K point mutation enhanced the invasion and migration abilities of ESCC cell lines
Through scratch experiments and Transwell experiments, the changes in the migration, invasion and metastasis of the KYSE140/450 cell lines under the effect of the NOTCH1 p.E450K point mutation were investigated. The results of the scratch experiment showed that the NOTCH1 p.E450K point mutation resulted in the enhanced invasion and migration abilities of KYSE140 and KYSE450 cells (Figure 4A,4B). Transwell experiments showed that the NOTCH1 p.E450K point mutation enhanced the invasiveness of the KYSE140 and KYSE450 cell lines (Figure 4C,4D).
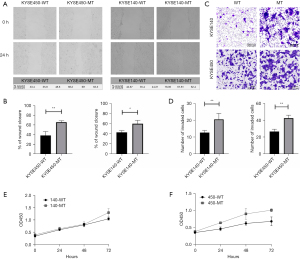
NOTCH1 p.E450K point mutation promoted proliferation
To figure out the effect of NOTCH1 p.E450K point mutation on the proliferation, we compared WT esophageal cancer cell lines with MT esophageal cancer cell lines using CCK-8 assay. There was no statistical significance in the comparison of proliferative ability of ESCC cells before and after NOTCH1 p.E450K point mutation, indicating that point mutation had no significant effect on the proliferation of ESCC cells (Figure 4E,4F).
The effects of the point mutation on the cell cycle
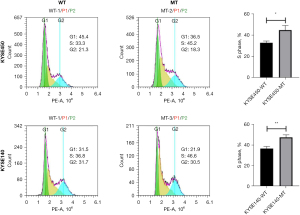
Flow cytometry analysis showed that the NOTCH1 p.E450K point mutation caused a significant increase in the proportion of KYSE140 and KYSE450 cells in S phase (Figure 5). S phase is the main period of DNA synthesis, also known as the DNA replication phase, during which the DNA number doubles. In general, once cells enter S phase, cell division continues until the G1 phase of the next cycle. An increase in cells in S phase indicates proliferation.
Discussion
Although it has been confirmed in many cancers, including esophageal cancer, that the activation of the NOTCH1 pathway leads to drug resistance in malignant tumors (7), little is known about the NOTCH pathway causing drug resistance in ESCC. In previous reports on ESCC gene sequencing, the frequency of NOTCH1 mutations was lower only than that of p53 (11,12). Many studies (13-22) have explained how the activation of the NOTCH1 signaling pathway leads to chemotherapy resistance. However, these studies mostly addressed the regulation of NOTCH1 transcription or expression, followed by the abnormal activation of the NOTCH1 pathway, which leads to chemotherapy resistance. No NOTCH1 sense mutations have been found, nor reports of NOTCH1 mutations that affect the ligand binding status of receptors in this pathway, resulting in chemotherapy resistance.
A previous study on drug resistance to nCT in esophageal cancer showed that in patients with ineffective chemotherapy, the detection rate of NOTCH1 missense mutations was significantly increased (9). Further study found that the p.E450K missense mutation increased the binding ability of the NOTCH1 receptor protein to its ligand DLL4 (10). On this basis, we investigated the effect of the NOTCH1 p.E450K missense mutation on cell phenotype to elucidate the biofunctional changes in NOTCH1 missense mutation cell lines. The results showed that the NOTCH1 p.E450K point mutation caused chemotherapy resistance in KYSE140 and KYSE450 ESCC cell lines, enhanced cell migration and invasion abilities, and increased the number of cells in S phase.
A study by Xie et al. (23) showed that high NOTCH1 expression promoted resistance to epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) in non-small cell lung cancer (NSCLC). Zhang et al. (24) showed that the NOTCH signaling pathway was activated in the erlotinib-resistant NSCLC cell line HCC827/ER. In a breast cancer study, Pandya et al. (25) found that positive human epidermal growth factor receptor 2 (ErbB-2) expression inhibited the transcriptional activity of NOTCH and that herceptin treatment activated the NOTCH signaling pathway and led to chemotherapy resistance. In the T47D model of tamoxifen-resistant breast cancer, abnormally activated PKCa specifically upregulated the expression of NOTCH to cause drug resistance in breast cancer cells (26). Hang et al. (27) found that NOTCH1 was highly expressed in cisplatin-resistant gastric cancer cell lines. Mirone et al. (28) found that NOTCH1 was significantly highly expressed in colorectal cancer (CRC) cell lines that were resistant to regorafenib and that the downregulation of NOTCH1 expression reversed drug resistance. Huang et al. (29) found that in CRC patients, NOTCH1 was abnormally highly expressed in cells resistant to oxaliplatin and fluorouracil. Takam et al. (30) found that NOTCH1 and the downstream target gene Hes1 of the NOTCH signaling pathway were highly expressed in human bone marrow mesenchymal stem cells (hBM-MSCs) from acute myeloid leukemia (AML) patients and that hBM-MSCs mediated and activated the NOTCH signaling pathway to promote the proliferation and drug resistance of leukemia cells. In chemotherapy sensitivity studies of breast cancer (31) and prostate cancer (32), it was found that the sensitivity of some patients to chemotherapeutic drugs could be improved by downregulating the NOTCH1 signaling pathway, suggesting that the NOTCH1 receptor and its ligand-mediated signaling pathways are closely related to chemotherapy resistance.
In the drug sensitivity test of the KYSE140 ESCC cell lines, the results showed that as the concentration of paclitaxel/cisplatin increased, its inhibitory effect on the KYSE140-WT and KYSE140-MT cell lines gradually increased (cell viability gradually decreased) in a dose-dependent manner. The IC50 results for paclitaxel/cisplatin showed that the IC50 concentration for the cells in the KYSE140-WT group was significantly lower than that for the cells in the KYSE140-MT group (P<0.01), indicating that signaling pathways in the KYSE140 cell line with the NOTCH missense mutation may be abnormally activated. Activation leads to the drug-resistant phenotype in tumor cells and the enhancement of chemotherapy resistance in ESCC cell lines. The same conclusion was obtained in the comparison of KYSE450-MT and KYSE450-WT cells.
The NOTCH signaling pathway is closely related to cell proliferation and differentiation and plays a very important regulatory role in body development. A number of studies have found that abnormal NOTCH signaling is associated with various common tumors, such as gastric cancer (33), breast cancer (34), lung cancer (35), and liver cancer (36). Some studies (37,38) also found that NOTCH signaling is involved in the process of tumor invasion and metastasis. A study on ESCC (7) explored the association between gene mutations in tumor cells and prognosis and chemotherapy response. The results showed that patients with NOTCH1 mutations survived for a shorter time than patients without NOTCH1 mutations and patients with NOTCH1 mutations did not respond to chemotherapy. By comparing pathological staging among patients with HCC, Wang et al. (39) found that NOTCH1 gene expression in patients with late-stage disease (stage III/IV) was higher than that in patients with earlier stage disease (stage I/II), suggesting that the NOTCH1 gene expression level may be associated with the invasion and metastasis of hepatocellular carcinoma (HCC). Another study (40) found that Jagged1, a NOTCH signaling ligand, was associated with the recurrence and metastasis of prostate cancer. Some studies (41,42) have confirmed that the NOTCH pathway can synergize with other tumor signaling pathways, such as the EGFR, Ras, and Akt signaling pathways, which are very important for tumor development.
In this study, in the scratch and Transwell experiments, the MT cell line had stronger invasion and migration abilities than the WT cell line (P<0.05). Flow cytometry analysis also showed that the proportion of cells in S phase among the MT cells was significantly higher than that among WT cells, and proliferation test results showed that the proliferation ability of NOTCH1 mutation cell line was enhanced. In general, once cells enter S phase, cell division continues until the G1 phase of the next cycle. An increase in the number of tumor cells in S phase indicates tumor cell proliferation and increases the ability of tumor cells to resist chemotherapy.
In the KYSE140 and KYSE450 ESCC cell lines, the NOTCH1 p.E450K point mutation enhanced the migration and invasion abilities and increased the percentage of cells in S phase. Although cell function experiments have yielded satisfactory results, they only reflect laboratory conditions, and therefore, further verification is needed by carrying out corresponding animal tumor model experiments. ESCC is a malignant tumor with high incidence and strong invasiveness. Although nCT has achieved good effects, resistance of tumor cells to chemotherapy is still a problem. If the mechanism underlying the involvement of the NOTCH signaling pathway in cell drug resistance is elucidated, it may provide new therapeutic targets and targeted pathways for molecular targeted therapy for the reversal of drug resistance in esophageal cancer. This missense mutation may be a potential predictor of clinical chemotherapy efficacy and an important target for the reversal of drug resistance, which is worthy of further study.
Conclusions
The NOTCH1 p.E450K point mutation causes chemotherapy resistance in KYSE140 and KYSE450 ESCC cells. Cell functional experiments showed that the NOTCH1 p.E450K point mutation enhanced the proliferation, migration and invasion abilities of KYSE140 and KYSE450 cells and increased the number of cells in S phase; the mutation had no effect on apoptosis.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1880/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1880/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1880/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1880/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was reviewed and approved by the Ethics Review Committee of the Affiliated Cancer Hospital of Zhengzhou University/Henan Cancer Hospital (No. 2021-KY-0049-001).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Arnold M, Soerjomataram I, Ferlay J, et al. Global incidence of oesophageal cancer by histological subtype in 2012. Gut 2015;64:381-7. [Crossref] [PubMed]
- Zheng Y, Li Y, Liu X, et al. Reevaluation of Neoadjuvant Chemotherapy for Esophageal Squamous Cell Carcinoma: A Meta-Analysis of Randomized Controlled Trials Over the Past 20 Years. Medicine (Baltimore) 2015;94:e1102. [Crossref] [PubMed]
- Wang E, Zaman N, Mcgee S, et al. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol 2015;30:4-12. [Crossref] [PubMed]
- Dang TP. Notch, apoptosis and cancer. Adv Exp Med Biol 2012;727:199-209. [Crossref] [PubMed]
- Hu YY, Zheng MH, Zhang R, et al. Notch signaling pathway and cancer metastasis. Adv Exp Med Biol 2012;727:186-98. [Crossref] [PubMed]
- Song B, Cui H, Li Y, et al. Mutually exclusive mutations in NOTCH1 and PIK3CA associated with clinical prognosis and chemotherapy responses of esophageal squamous cell carcinoma in China. Oncotarget 2016;7:3599-613. [Crossref] [PubMed]
- Zhang H, Jiang H, Chen L, et al. Inhibition of Notch1/Hes1 signaling pathway improves radiosensitivity of colorectal cancer cells. Eur J Pharmacol 2018;818:364-70. [Crossref] [PubMed]
- Liu J, Xing W, Tian Q, et al. Application of next-generation sequencing in resistance genes of neoadjuvant chemotherapy for esophageal cancer. Transl Cancer Res 2020;9:4847-56. [Crossref] [PubMed]
- Gao K, Xing W, Liu X, et al. The Notch1 gene may control cell chemoresistance in esophageal squamous cell cancer. Transl Cancer Res 2021;10:3278-85. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015;96:597-611. [Crossref] [PubMed]
- Alcolea MP, Jones PH. Cell competition: winning out by losing notch. Cell Cycle 2015;14:9-17. [Crossref] [PubMed]
- Kang M, Jiang B, Xu B, et al. Delta like ligand 4 induces impaired chemo-drug delivery and enhanced chemoresistance in pancreatic cancer. Cancer Lett 2013;330:11-21. [Crossref] [PubMed]
- Cenciarelli C, Marei HE, Zonfrillo M, et al. The interference of Notch1 target Hes1 affects cell growth, differentiation and invasiveness of glioblastoma stem cells through modulation of multiple oncogenic targets. Oncotarget 2017;8:17873-86. [Crossref] [PubMed]
- Lee CW, Raskett CM, Prudovsky I, et al. Molecular dependence of estrogen receptor-negative breast cancer on a notch-survivin signaling axis. Cancer Res 2008;68:5273-81. [Crossref] [PubMed]
- Roodhart JM, He H, Daenen LG, et al. Notch1 regulates angio-supportive bone marrow-derived cells in mice: relevance to chemoresistance. Blood 2013;122:143-53. [Crossref] [PubMed]
- Zhang Y, Xu W, Guo H, et al. NOTCH1 Signaling Regulates Self-Renewal and Platinum Chemoresistance of Cancer Stem-like Cells in Human Non-Small Cell Lung Cancer. Cancer Res 2017;77:3082-91. [Crossref] [PubMed]
- Chen Y, Li D, Liu H, et al. Notch-1 signaling facilitates survivin expression in human non-small cell lung cancer cells. Cancer Biol Ther 2011;11:14-21. [Crossref] [PubMed]
- Katoh M, Katoh M. Notch signaling in gastrointestinal tract Int J Oncol 2007;30:247-51. (review).
- Joshi I, Minter LM, Telfer J, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 2009;113:1689-98. [Crossref] [PubMed]
- Sun B, Dong C, Lei H, et al. Propranolol inhibits proliferation and invasion of hemangioma-derived endothelial cells by suppressing the DLL4/Notch1/Akt pathway. Chem Biol Interact 2018;294:28-33. [Crossref] [PubMed]
- Xie M, He CS, Wei SH, et al. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur J Cancer 2013;49:3559-72. [Crossref] [PubMed]
- Zhang H, Chen F, He Y, et al. Sensitivity of non-small cell lung cancer to erlotinib is regulated by the Notch/miR-223/FBXW7 pathway. Biosci Rep 2017;37:BSR20160478. [Crossref] [PubMed]
- Pandya K, Wyatt D, Gallagher B, et al. PKCα Attenuates Jagged-1-Mediated Notch Signaling in ErbB-2-Positive Breast Cancer to Reverse Trastuzumab Resistance. Clin Cancer Res 2016;22:175-86. [Crossref] [PubMed]
- Yun J, Pannuti A, Espinoza I, et al. Crosstalk between PKCα and Notch-4 in endocrine-resistant breast cancer cells. Oncogenesis 2013;2:e60. [Crossref] [PubMed]
- Hang Q, Sun R, Jiang C, et al. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anticancer Drugs 2015;26:632-40. [Crossref] [PubMed]
- Mirone G, Perna S, Shukla A, et al. Involvement of Notch-1 in Resistance to Regorafenib in Colon Cancer Cells. J Cell Physiol 2016;231:1097-105. [Crossref] [PubMed]
- Huang R, Wang G, Song Y, et al. Colorectal cancer stem cell and chemoresistant colorectal cancer cell phenotypes and increased sensitivity to Notch pathway inhibitor. Mol Med Rep 2015;12:2417-24. [Crossref] [PubMed]
- Takam Kamga P, Bassi G, Cassaro A, et al. Notch signalling drives bone marrow stromal cell-mediated chemoresistance in acute myeloid leukemia. Oncotarget 2016;7:21713-27. [Crossref] [PubMed]
- Kang L, Mao J, Tao Y, et al. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci 2015;106:700-8. [Crossref] [PubMed]
- Liu X, Luo X, Wu Y, et al. MicroRNA-34a Attenuates Paclitaxel Resistance in Prostate Cancer Cells via Direct Suppression of JAG1/Notch1 Axis. Cell Physiol Biochem 2018;50:261-76. [Crossref] [PubMed]
- Li GG, Li L, Li C, et al. Influence of up-regulation of Notch ligand DLL4 on biological behaviors of human gastric cancer cells. World J Gastroenterol 2013;19:4486-94. [Crossref] [PubMed]
- Mittal S, Subramanyam D, Dey D, et al. Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis. Mol Cancer 2009;8:128. [Crossref] [PubMed]
- Xu K, Moghal N, Egan SE. Notch signaling in lung development and disease. Adv Exp Med Biol 2012;727:89-98. [Crossref] [PubMed]
- Villanueva A, Alsinet C, Yanger K, et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 2012;143:1660-1669.e7. [Crossref] [PubMed]
- Bolós V, Mira E, Martínez-Poveda B, et al. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res 2013;15:R54. [Crossref] [PubMed]
- Chen J, Imanaka N, Chen J, et al. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer 2010;102:351-60. [Crossref] [PubMed]
- Wang XQ, Zhang W, Lui EL, et al. Notch1-Snail1-E-cadherin pathway in metastatic hepatocellular carcinoma. Int J Cancer 2012;131:E163-72. [Crossref] [PubMed]
- Santagata S, Demichelis F, Riva A, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res 2004;64:6854-7. [Crossref] [PubMed]
- Osipo C, Golde TE, Osborne BA, et al. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest 2008;88:11-7. [Crossref] [PubMed]
- Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev 2005;19:1825-39. [Crossref] [PubMed]


