
Molecular characteristics and multivariate survival analysis of 43 patients with locally advanced or metastatic esophageal squamous cell carcinoma
Highlight box
Key findings
• The mutational landscape of 43 Chinese esophageal squamous cell carcinoma (ESCC) patients was revealed, and the correlations between tumor mutation burden (TMB), the gene mutations of NOTCH1, CBLB, and TSC2, and overall survival (OS) were identified.
What is known and what is new?
• NOTCH1, CBLB and TSC2 mutations were significantly associated with poor OS, consistent with the results of previous studies.
• Patients with TMB ≥7 had significantly longer OS.
What is the implication, and what should change now?
• TMB was found to be associated with OS, which could potentially guide the treatment and management of patients with ESCC. To validate and expand upon our results, larger studies and extensive clinical follow-ups will be needed in the future.
Introduction
Esophageal cancer (EC) is an aggressive malignant tumor with poor prognosis and high incidence. It is the sixth leading cause of cancer-related death in the world (1). About 35% of all patients with EC are in advanced stages at the time of presentation, so these patients lose the opportunity to receive radical treatment, which does not respond well, with an overall 5-year survival rate ranging from 12% to 20% (2-4). Esophageal squamous cell carcinoma (ESCC) occurs primarily in Asian populations, while esophageal adenocarcinoma (EAC) is more common in North America and Western Europe (1,5). Smoking and drinking alcohol are two risk factors for ESCC (6), whereas obesity is a risk factor for EAC (7).
Surgery is the primary treatment for resectable EC without distant metastasis. In patients with locally advanced EC, the R0 excision rate is low (about 50%), leading to early postoperative recurrence and thus long-term survival remains low (8). Targeted therapeutic approaches have also been investigated in EC patients, including human epidermal growth factor receptor 2 (HER2)-targeted therapy, anti-angiogenic therapy, and immunotherapy. Anti-HER2 monoclonal antibody trastuzumab has been approved by the U.S. Food and Drug Administration (FDA) for use in patients with advanced HER2-positive EC (9). Ramucirumab, a VEGF receptor 2 (VEGFR-2) antibody, has received approval for the treatment of patients with advanced or metastatic EAC, initially as monotherapy and subsequently in combination with paclitaxel (10). In addition, pembrolizumab and camrelizumab have been shown to improve survival better than chemotherapy in second-line treatment of patients with advanced or metastatic EC (11,12).
Recent advancements in next-generation sequencing (NGS) studies have enhanced our understanding of genetic alterations in EC genes (13). With the help of NGS technologies, molecular characterization is emerging as a new aspect of multidisciplinary treatment decision making to develop holistic treatment plans for cancer patients (14). Genes involved in the cell cycle, the RTK/PI3K/AKT circuit, chromatin remodeling, and the NOTCH signaling pathway are frequently altered (13). Genes with a high frequency of mutations in EC, including TP53, NOTCH1, PIK3CA, CDKN2A and RB1, have been identified and reported in different races (15,16), in which TP53 is the most frequently mutated gene in ESCC, exhibiting a mutation frequency of 93% (13). Associations between these genetic alterations and prognosis have been reported. In our study, we enrolled 43 patients with locally advanced or metastatic ESCC, collected the clinical data of these patients, and conducted NGS detection on their tumor tissues and blood samples using 425 gene-panel. The aim of this study was to analyze the molecular characteristics and explore the relationship between genetic alterations and prognosis of ESCC patients. Further studies on the correlation between abnormalities in related signaling pathways and survival provide a theoretical basis for the subsequent extension of the survival of patients with ESCC. We present this article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1601/rc).
Methods
Patient cohort
According to the National Comprehensive Cancer Network (NCCN) guidelines, 43 non-surgical patients with locally advanced or metastatic ESCC were retrospectively recruited from Zhejiang Cancer Hospital. The inclusion criteria were as follows: (I) all patients were histologically confirmed to have primary nonoperative ESCC, (II) patients underwent treatment with either chemoradiotherapy or immunotherapy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Review Committee of Zhejiang Cancer Hospital (IRB-2018-20) and informed written consent was obtained from each participant.
DNA extraction and library construction
Genomic DNA was prepared using the QIAamp DNA FFPE tissue Kit (Qiagen, Hilden, Germany) and the QIAamp DNA Blood Mini kit (Qiagen). The amount of extracted DNA was assessed using a Qubit 3.0 fluorometer and the quality of DNA was measured using a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, USA). The library for sequencing was prepared with the KAPA Hyper Prep kit from KAPA Biosystems and was sequenced using a targeted panel containing 425 cancer-related genes.
Bioinformatics analysis
The results of high-throughput sequencing were compared with the genome data of Chinese hG19 to complete gene mapping. Multiple gene mutations, such as gene point mutation, insertion deletion mutation, gene copy number change and chromosome structure abnormality, were analyzed synchronously from the mapping results. Furthermore, mutations were analyzed and generated by comparison with 1000 Genome database and dbSNP database. We estimated each patient’s tumor mutation burden (TMB) by counting the somatic mutations in coding regions, including single-nucleotide mutations and insertions or deletions (indels), without counting driver mutations and known germline changes.
Statistical analysis
Kaplan-Meier survival analysis was used to draw survival curve and evaluate overall survival (OS). Cox proportional risk model was used to analyze the relationship between mutation gene and prognosis. In addition to age and sex, non-significant confounding factors in univariate Cox model were excluded from the multivariate Cox analysis. Kaplan-Meier survival and Cox regression analysis were executed using R package (R 3.6.0). A P value less than 0.05 was considered statistically significant. Medication information for the ESCC patients was not used.
Results
Description of patient cohort
The clinicopathological features of 43 Chinese patients with nonsurgical ESCC are shown in Table 1. The median age of these patients was 61 years, ranging from 43 to 81 years. More than 80% (86.0%) of the patients were male and the remainders (14.0%) were female. All patients had squamous cell carcinoma (SCC, 100%). There were 1 patient (2.3%) in stage IIIA, 12 patients (27.9%) in stage IIIB and 30 patients (69.8%) in stage IV. Among them, 32 cases (74.4%) were smokers, and more than half (72.1%) had drinking history. All patients received radiotherapy, chemotherapy or immunotherapy.
Table 1
| Characteristics | Patients (%) |
|---|---|
| Age (years) | |
| <60 | 19 (44.2) |
| ≥60 | 24 (55.8) |
| Median [range] | 61 [43–81] |
| Sex | |
| Male | 37 (86.0) |
| Female | 6 (14.0) |
| Clinical stage | |
| IIIA | 1 (2.3) |
| IIIB | 12 (27.9) |
| IV | 30 (69.8) |
| Pathological subtype | |
| Squamous cell carcinoma | 43 (100.0) |
| Adenocarcinoma | 0 (0) |
| Metastasis | |
| Yes | 38 (88.4) |
| No | 5 (11.6) |
| Smoking status | |
| Never-smoker | 11 (25.6) |
| Former-smoker | 32 (74.4) |
| Drinking status | |
| No alcohol consumption | 12 (27.9) |
| Alcohol consumption | 31 (72.1) |
| Tumor location | |
| Upper thoracic | 10 (23.3) |
| Middle thoracic | 32 (74.4) |
| Lower thoracic | 1 (2.3) |
Genetic mapping of Chinese patients with ESCC
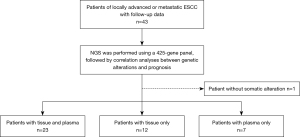
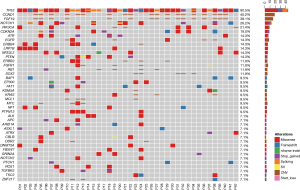
We obtained 35 tumor tissue samples and 31 blood samples from 43 Chinese non-surgical ESCC patients, of which 23 patients had both tissue and blood samples, 12 patients had tissue samples only, and eight patients had only tissue samples. Blood samples, since only one germline mutation was detected in one patient, were described only and not included in subsequent survival analysis (Figure 1). The somatic gene mutation maps of the other 42 patients with ESCC were shown in Figure 2. The most commonly mutated genes were TP53 (90.5%), CCND1 (45.2%), FGF19 (38.1%), NOTCH1 (26.2%), PI3KCA (21.4%) and CDKN2A (19%). The most common gene amplifications were CCND1, FGF19. Among these mutations, PI3KCA and NOTCH1 exhibited a certain degree of mutual exclusion, which is consistent with previous reports (17).
Univariate and multivariate Cox regression analysis of prognostic factors
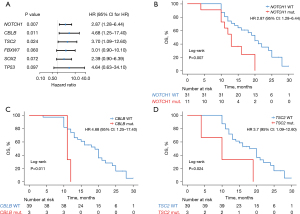
Genetic mutations in patients with EC can be a potential predictor of prognosis during cancer treatment. In this study, 43 patients with ESCC had a median OS of 16 months (Figure S1). We used univariate Cox regression model to investigate the relationship between genes with high mutation frequency and OS. Based on previous reports, we conducted univariate analysis of the relationship between NOTCH1, CBLB, TSC2, FBXW7, SOX2, TP53 and OS of these patients. As shown in Figure 3, NOTCH1 (P=0.007, HR =2.87, Figure 3B), CBLB (P=0.011, HR =4.68, Figure 3C) and TSC2 (P=0.024, HR =3.7, Figure 3D) were significantly associated with poorer OS. TP53, which exhibited the highest mutation frequency, could not predict OS (P=0.097, HR =4.64, Figure 3A), which may be due to the limited number of patients with wild-type TP53.
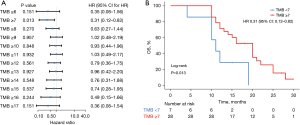
Later, we conducted a univariate analysis of 35 patients with tissue TMB (Figure 4A), and the results showed that when TMB =7 was divided as the boundary, patients with TMB ≥7 had longer OS, HR =0.31, and P value was significant (Figure 4B). When TMB ranged from 14 to 17, patients with TMB-H showed a trend of better OS, but there was no statistical difference (Figure 4A).
In the above univariate model, NOTCH1, CBLB, TSC2 gene mutations and TMB ≥7 were independent markers of OS (Table 2). Multivariate Cox regression analysis was performed for these four factors. Multivariate analysis showed that CBLB was an independent factor, and patients with CBLB gene mutation had worse OS (HR =5.24, P=0.02), which was significant and is consistent with previously reported results (16).
Table 2
| Variable | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| NOTCH1 mutation | 2.87 (1.28–6.44) | 0.007 | 2.31 (0.93–5.79) | 0.073 | |
| CBLB mutation | 4.68 (1.25–17.40) | 0.011 | 5.24 (1.30–21.12) | 0.020 | |
| TSC2 mutation | 3.70 (1.09–12.60) | 0.024 | 2.54 (0.52–12.30) | 0.248 | |
| TMB ≥7 | 0.31 (0.116–0.82) | 0.013 | 0.42 (0.12–1.47) | 0.173 | |
TMB, tumor mutation burden; OS, overall survival; HR, hazard ratio; CI, confidence interval.
Alterations in signaling pathways and survival of EC patients
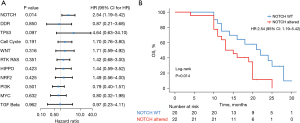
NOTCH signaling is a contact-dependent cell-cell communication event in multicellular organisms and plays a fundamental role in embryonic development, differentiation, proliferation and apoptosis, and regulation of cell fate (18,19). NOTCH signaling is frequently altered in EC (20). Based on the fact that NOTCH1 is a common mutant gene in the above studies, we conducted correlation analysis between alterations in NOTCH signaling pathway and OS in ESCC patients. Results as shown in Figure 5, in this study, the OS of ESCC patients with NOTCH signaling pathway abnormalities was significantly worse, HR =2.54, P=0.014. Previous literature has shown that abnormal NOTCH signaling pathway are associated with EC development and poor prognosis, which is consistent with our results (21,22). Meanwhile, we also studied the relationship between genetic alterations in DNA damage repair (DDR) and other important signaling pathways and OS, but the results were not significant (Figure S2).
Discussion
EC is one of the most common cancers in China, accounting for half of global cases (23). EC is primarily composed of SCC or adenocarcinoma (AC), which are associated with anatomical location, natural history, environmental risk factors, and treatment outcome (24). ESCC mainly originates in the upper two-thirds of the esophagus, and long-term exposure to tobacco and alcohol increases the risk. Conversely, EAC tends to be distal to the esophagus, closed to the gastroesophageal junction (GEJ), and is associated with long-term acid reflux and Barrett’s esophagus (25). Histopathologically, SCC progresses from epithelial dysplasia to carcinoma in situ, and eventually to invasive disease (26). AC appears in the background of intestinal metaplasia, and its growth pattern is similar to that of Lauren enteric stomach AC.
Based on the development of NGS, more and more somatic mutations in EC genomes have been reported (27). The mutation frequency of TP53, NOTCH1, NOTCH2, NOTCH3, FBXW7, MYCBP2, KIF16B and KIF21B is higher in the North American and Western European populations, while in the Chinese population, the mutation frequency of TP53, PIK3CA, NOTCH1, FAT1, FAT2, ZNF750 and KMT2D is high (14,28). In this study, we analyzed the molecular characteristics of 43 ESCC patients in China using a pan-carcinomatous NGS panel containing 425 genes. High frequencies of TP53, CCND1, FGF19, CDKN2A, PI3KCA and NOTCH1 alterations were detected. Meanwhile, we found that TP53, CCND1, FGF19, CDKN2A and PI3KCA high-frequency mutations were not associated with OS, while NOTCH1, CBLB and TSC2 were significantly associated with poor OS, consistent with results from previous studies (16,29).
Based on our results, we identified the alteration of NOTCH1, CBLB and TSC2 genes as adverse prognostic factors. The NOTCH gene was firstly identified in fruit flies in 1919 and gets its name from the fact that it produces NOTCH on the edge of the fly’s wing (30). NOTCH receptors are mainly composed of intracellular, transmembrane and extracellular domains and are expressed as heterodimer proteins on the cell surface. NOTCH intracellular domain (NICD) consists of five parts: (I) the RAM region, which binds DNA binding proteins; (II) six anchor protein repeats (ANK), as enhancers of NOTCH, mediate the interaction between NOTCH and other proteins; (III) two nuclear localization signals (NLS); (IV) trans-activation domains (TAD) that have not been defined for NOTCH3 and NOTCH4; (V) a proline-glutamate-serine-threonine domain (PEST) region associated with the stability of NOTCH receptor (31). The extracellular domain includes 29–36 epidermal growth factor (EGF) -like repeats and three cysteine Lin NOTCH repeats (LNRs). The main role of these regions is to bind ligands and initiate NOTCH (31). NOTCH signaling pathway controls differentiation, proliferation and apoptosis in mammalian tissues. NOTCH signaling abnormalities, including gain of function (GoF) of NOTCH1 alteration and loss of function (LoF) of NOTCH1 alteration, are involved in the biological processes of multiple tumors, such as GoF of NOTCH1 alteration, which has been found in chronic lymphocytic leukemia, T-cell acute lymphoblastic leukemia (T-ALL), mantle cell lymphoma, diffuse large B-cell lymphoma, breast cancer and non-small cell lung cancer (NSCLC) (32-37), while LoF NOTCH1 has been found in tumors such as head and neck SCC (HNSCC), cutaneous SCC (CSCC), and small cell lung cancer (SCLC) (38-40). The role of NOTCH signaling pathway in ESCC has not been studied until recently. A study of exome sequencing in 12 ESCC patients showed that NOTCH1 inactivation mutations were present in 21% of patients (28). Study by Martincorena et al. has shown that NOTCH1 mutation occurred with much greater frequency in normal oesophagus compared to EC (41). In another study, Wang et al. investigated the invasion and metastasis of EC cell line EC-9706 by inducing NOTCH1-mediated epithelial-mesenchymal transformation (EMT) in snail. Results showed that inhibition of NOTCH1 expression in EC cell line EC-9706 inhibited EMT. It also diminished the capacity for invasion and metastasis of EC-9706 cells, suggesting that NOTCH1 was involved in the invasion and spread of EC through snail mediated EMT, and inhibition of NOTCH1 signaling pathway can inhibit the invasion and metastasis of tumor, suggesting that NOTCH1 may be an effective target for the prevention and treatment of EC (42). This suggests that NOTCH1 signaling has an intrinsic tumorigenic effect. In this study, we conducted a correlation analysis between abnormalities in NOTCH signaling pathway and OS in ESCC patients, and found that the alterations in NOTCH signaling pathway had poor OS in ESCC patients, suggesting that abnormal NOTCH signaling pathway are related to the development of ESCC and may have an intrinsic tumorigenic effect, which is consistent with previous reports. However, due to different research methods and small sample size, the complexity of NOTCH1 signaling pathway in ESCC still needs to be explored for a long time.
Ubiquitination is a post-translational modification that targets cellular proteins for degradation (43). CBLB, a second member of the E3 ubiquitin ligase CBL family, has been found to be involved in ubiquitination regulation of cell proliferation, migration, and drug sensitivity (44). It has been reported that CBLB regulates multidrug resistance of human gastric cancer cells through different signaling pathways (45). In another study, Chen et al. found that Mir-27B-3p inhibits proliferation of breast cancer cells by targeting CBLB/GRB2 and may reverse multidrug resistance (46). However, the role of CBLB in chemotherapy resistance of EC has been neglected. Recently, Yang et al. conducted in vitro experiments on cisplatin-resistant EAC cells OE19/CDDP and parental sensitive OE19 cells, and found that CBLB was a direct target of Mir-181A-5p and was reverse-regulated by Mir-181A-5p. Compared with OE19 cells, CBLB was highly expressed in OE19/CDDP cells, and the overexpression of CBLB aggravated the resistance of EAC cells to cisplatin (47). These results suggest that CBLB is a drug-resistant gene associated with cisplatin therapy for EC. In this study, univariate and multivariate Cox regression analysis showed that CBLB mutation was associated with poor prognosis in patients with ESCC. Multivariate analysis showed that CBLB was an independent factor, and patients with CBLB gene mutation had worse OS. TSC2 acts as a negative regulator upstream of mTOR, and the TSC2-mTOR signaling pathway is crucial in the regulation of tumor autophagy (48). TSC2 inactivation mutations can lead to tuberous sclerosis complex, an autosomal dominant syndrome that can lead to the development of multi-organ tumors. TSC1, TSC2 and TBC1D7 together form the TSC protein complex, which acts as a GAP of the GTPase RHEB and inhibits rapamycin complex-1 (mTORC1) (48). In a previous study, Chen et al. found that active TSC2 inhibits mTOR through RHEB by detecting differential phosphorylation of short TSC2 or long TSC2 splicing variants and its effect on mTOR signaling pathway, leading to continuous stimulation of carcinogenic autophagy in ESCC cells (49). These results suggest that TSC2 gene is related to the occurrence and development of tumor. In this study, TSC2 mutation was found to be significantly associated with shorter OS in ESCC patients, which is consistent with previous reports. TMB has been identified as a novel predictive biomarker for the efficacy of immune checkpoint blockade (ICB). Higher TMB is usually associated with better OS after ICB treatment in multiple cancers, including NSCLC, bladder cancer, colorectal cancer and melanoma (50). Programmed death-ligand 1 (PD-L1) expression in EC immunotherapy cannot accurately predict the efficacy of PD-1 inhibitors in EC patients (51). On this basis, TMB has gradually become a biomarker for the immunotherapy of EC. In patients who received a combination of nivolumab and ipilimumab, and standard of care (SOC) chemotherapy, a therapeutic benefit dependent on TMB but not PD-L1 expression levels was observed (52). This condition is thought to exist in tumors that have a high TMB, T cell infiltration and/or activation regulated in a CTLA-4 dependent manner (53). Pang et al.’s study on TMB may indicate that high-risk EC patients are more responsive to immunotherapy due to high TMB (54). In our study, we also conducted a univariate analysis of 35 patients with tumor TMB, and the results showed that patients with TMB ≥7 had significantly longer OS. However, when the threshold ranged from 14 to 17, patients with TMB-H only showed a trend towards better OS, but the difference was not statistically significant. This is different from the high TMB reported previously and may be related to the small sample size and clinical follow-up. Nonetheless, this result could offer a more comprehensive perspective for our subsequent research on TMB in predicting the immune efficacy of ESCC, not just confined to high TMB.
Conclusions
We studied the mutational landscape of 43 Chinese patients with locally advanced or metastatic ESCC and discovered the correlation between TMB and OS. We further explored the relationship between NOTCH, DDR and other important signaling pathway abnormalities and OS, as well as the potential prognostic relationship of NOTCH1, CBLB and TSC2 mutations. However, the limitation of this study lies in its small sample size and the difficulty in clinical follow-up. Larger studies and more extensive clinical follow-up are needed to confirm and extend our findings in the future.
Acknowledgments
The authors would like to express their sincere thanks to all the patients and researchers involved.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1601/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1601/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1601/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1601/coif). D.W., Y.Z. and J.Y. are employees of Nanjing Geneseeq Technology, Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Review Committee of Zhejiang Cancer Hospital (IRB-2018-20) and informed written consent was obtained from each participant.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol 2013;19:5598-606. [Crossref] [PubMed]
- Napier KJ, Scheerer M, Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol 2014;6:112-20. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. [Crossref] [PubMed]
- Turati F, Tramacere I, La Vecchia C, et al. A meta-analysis of body mass index and esophageal and gastric cardia adenocarcinoma. Ann Oncol 2013;24:609-17. [Crossref] [PubMed]
- Ajani JA, D’Amico TA, Bentrem DJ, et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019;17:855-83. [Crossref] [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Shimodaira Y, Elimova E, Wadhwa R, et al. Ramucirumab for the treatment of gastroesophageal cancers. Expert Opin Orphan Drugs 2015;3:737-46. [Crossref] [PubMed]
- Huang J, Xu J, Chen Y, et al. Camrelizumab versus investigator’s choice of chemotherapy as second-line therapy for advanced or metastatic oesophageal squamous cell carcinoma (ESCORT): a multicentre, randomised, open-label, phase 3 study. Lancet Oncol 2020;21:832-42. [Crossref] [PubMed]
- Kojima T, Shah MA, Muro K, et al. Randomized Phase III KEYNOTE-181 Study of Pembrolizumab Versus Chemotherapy in Advanced Esophageal Cancer. J Clin Oncol 2020;38:4138-48. [Crossref] [PubMed]
- Sawada G, Niida A, Uchi R, et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology 2016;150:1171-82. [Crossref] [PubMed]
- Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91-5. [Crossref] [PubMed]
- Wang Y, Liang N, Xue Z, et al. Identifying an Eight-Gene Signature to Optimize Overall Survival Prediction of Esophageal Adenocarcinoma Using Bioinformatics Analysis of ceRNA Network. Onco Targets Ther 2020;13:13041-54. [Crossref] [PubMed]
- Zhang N, Shi J, Shi X, et al. Mutational Characterization and Potential Prognostic Biomarkers of Chinese Patients with Esophageal Squamous Cell Carcinoma. Onco Targets Ther 2020;13:12797-809. [Crossref] [PubMed]
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med 2014;371:2499-509. [Crossref] [PubMed]
- Chiba S. Notch signaling in stem cell systems. Stem Cells 2006;24:2437-47. [Crossref] [PubMed]
- Wang Z, Li Y, Banerjee S, et al. Emerging role of Notch in stem cells and cancer. Cancer Lett 2009;279:8-12. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-337.e10. [Crossref] [PubMed]
- Forghanifard MM, Taleb S, Abbaszadegan MR. Notch Signaling Target Genes are Directly Correlated to Esophageal Squamous Cell Carcinoma Tumorigenesis. Pathol Oncol Res 2015;21:463-7. [Crossref] [PubMed]
- Cheng C, Cui H, Zhang L, et al. Genomic analyses reveal FAM84B and the NOTCH pathway are associated with the progression of esophageal squamous cell carcinoma. Gigascience 2016;5:1. [Crossref] [PubMed]
- Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970-90. Int J Cancer 2002;102:271-4. [Crossref] [PubMed]
- Siewert JR, Stein HJ, Feith M, et al. Histologic tumor type is an independent prognostic parameter in esophageal cancer: lessons from more than 1,000 consecutive resections at a single center in the Western world. Ann Surg 2001;234:360-7; discussion 368-9. [Crossref] [PubMed]
- Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol 2007;17:38-44. [Crossref] [PubMed]
- Kuwano H, Saeki H, Kawaguchi H, et al. Proliferative activity of cancer cells in front and center areas of carcinoma in situ and invasive sites of esophageal squamous-cell carcinoma. Int J Cancer 1998;78:149-52. [Crossref] [PubMed]
- Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014;46:467-73. [Crossref] [PubMed]
- Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2012;2:899-905. [Crossref] [PubMed]
- Dai H, Shao YW, Tong X, et al. YAP1 amplification as a prognostic factor of definitive chemoradiotherapy in nonsurgical esophageal squamous cell carcinoma. Cancer Med 2020;9:1628-37. [Crossref] [PubMed]
- Kojika S, Griffin JD. Notch receptors and hematopoiesis. Exp Hematol 2001;29:1041-52. [Crossref] [PubMed]
- Radtke F, Schweisguth F, Pear W. The Notch ‘gospel’. EMBO Rep 2005;6:1120-5. [Crossref] [PubMed]
- Westhoff B, Colaluca IN, D’Ario G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A 2009;106:22293-8. [Crossref] [PubMed]
- Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 2012;119:1963-71. [Crossref] [PubMed]
- Wang K, Zhang Q, Li D, et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a γ-secretase inhibitor. Clin Cancer Res 2015;21:1487-96. [Crossref] [PubMed]
- Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016;16:145-62. [Crossref] [PubMed]
- De Bie J, Demeyer S, Alberti-Servera L, et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia 2018;32:1358-69. [Crossref] [PubMed]
- Karube K, Enjuanes A, Dlouhy I, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 2018;32:675-84. [Crossref] [PubMed]
- Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333:1154-7. [Crossref] [PubMed]
- Wang NJ, Sanborn Z, Arnett KL, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A 2011;108:17761-6. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Martincorena I, Fowler JC, Wabik A, et al. Somatic mutant clones colonize the human esophagus with age. Science 2018;362:911-7. [Crossref] [PubMed]
- Wang T, Xuan X, Pian L, et al. Notch-1-mediated esophageal carcinoma EC-9706 cell invasion and metastasis by inducing epithelial-mesenchymal transition through Snail. Tumour Biol 2014;35:1193-201. [Crossref] [PubMed]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002;82:373-428. [Crossref] [PubMed]
- Yingchun L, Xiujuan Q, Jinglei Q, et al. E3 ubiquitin ligase Cbl-b potentiates the apoptotic action of arsenic trioxide by inhibiting the PI3K/Akt pathway. Braz J Med Biol Res 2011;44:105-11. [Crossref] [PubMed]
- Yu P, Fan Y, Qu X, et al. Cbl-b regulates the sensitivity of cetuximab through ubiquitin-proteasome system in human gastric cancer cells. J BUON 2016;21:867-73. [PubMed]
- Chen D, Si W, Shen J, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis 2018;9:188. [Crossref] [PubMed]
- Yang S, Wang P, Wang S, et al. miRNA-181a-5p Enhances the Sensitivity of Cells to Cisplatin in Esophageal Adenocarcinoma by Targeting CBLB. Cancer Manag Res 2020;12:4981-90. [Crossref] [PubMed]
- Ranek MJ, Kokkonen-Simon KM, Chen A, et al. PKG1-modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature 2019;566:264-9. [Crossref] [PubMed]
- Chen Y, Lu Y, Ren Y, et al. Starvation-induced suppression of DAZAP1 by miR-10b integrates splicing control into TSC2-regulated oncogenic autophagy in esophageal squamous cell carcinoma. Theranostics 2020;10:4983-96. [Crossref] [PubMed]
- Bader JE, Voss K, Rathmell JC. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol Cell 2020;78:1019-33. [Crossref] [PubMed]
- Yang H, Wang K, Wang T, et al. The Combination Options and Predictive Biomarkers of PD-1/PD-L1 Inhibitors in Esophageal Cancer. Front Oncol 2020;10:300. [Crossref] [PubMed]
- Reck M, Schenker M, Lee KH, et al. Nivolumab plus ipilimumab versus chemotherapy as first-line treatment in advanced non-small-cell lung cancer with high tumour mutational burden: patient-reported outcomes results from the randomised, open-label, phase III CheckMate 227 trial. Eur J Cancer 2019;116:137-47. [Crossref] [PubMed]
- Hu H, She L, Liao M, et al. Cost-Effectiveness Analysis of Nivolumab Plus Ipilimumab vs. Chemotherapy as First-Line Therapy in Advanced Non-Small Cell Lung Cancer. Front Oncol 2020;10:1649. [Crossref] [PubMed]
- Pang J, Pan H, Yang C, et al. Prognostic Value of Immune-Related Multi-IncRNA Signatures Associated With Tumor Microenvironment in Esophageal Cancer. Front Genet 2021;12:722601. [Crossref] [PubMed]


