
A comparison of machine learning methods for radiomics modeling in prediction of occult lymph node metastasis in clinical stage IA lung adenocarcinoma patients
Highlight box
Key findings
• Extreme gradient boosting (XGB) is the best algorithm to build radiomics model for predicting occult lymph node metastasis (ONM) in patients with clinical stage IA lung adenocarcinoma, yielding an area under the curve (AUC) of 0.917, and the combined model incorporating radiomics features and clinical-radiological features can better predict ONM with a superior AUC of 0.933.
What is known and what is new?
• Radiomics extracts quantitative features from images, and ONM is a common and prognostic factor in stage IA lung adenocarcinoma.
• A comparison of seven algorithms for radiomics modeling was carried out, and a novel combined model with improved performance was constructed.
What is the implication, and what should change now?
• Radiomics modeling can be enhanced by XGB and by combining features. Further validation and accessibility considerations are warranted.
Introduction
High-resolution computed tomography (HRCT) and lung cancer screening have become more widespread, and an increasing number of peripheral small lung adenocarcinomas have been detected (1,2). It is reported that about 17% clinical IA patients has been proven to have pathological lymph node metastasis (LNM) after surgical resection (3). Notably, these clinical IA cases differ from pathological IA lung adenocarcinomas, which typically lack LNM. The condition that preoperative evaluation did not detect LNM but postoperative pathology confirmed LNM is called occult lymph node metastasis (ONM) (4). To ensure sufficient resection of lymph node (LN) lesions, systematic LN dissection is still recommended as standard method for early-stage lung adenocarcinoma patients. Which means that more than 80% of clinical stage IA lung cancer patients have undergone unnecessary LN dissection. Invalid LN dissection may lead to prolonged postoperative recovery and increased the possibility of postoperative complications (5-7). Therefore, if ONM can be accurately diagnosed before surgery, targeted LN dissection can be performed on clinical stage IA lung adenocarcinoma patients to reduce unnecessary surgical damage.
Nevertheless, the accurate diagnosis of LN status in early lung adenocarcinoma patients remains a significant challenge. Studies have investigated the correlation between radiological features of the primary tumor and LNM in lung adenocarcinoma (8-10). Certain visual signs observed on computed tomography (CT) images, such as the solid component size, bronchial cutoff sign, and spiculation sign, have been identified as predictive factors for LNM. However, these features are inherently subjective and cannot be quantified. In addition, a vast amount of invisible information underlying radiographic images cannot be visually inspected or utilized. Radiomics, an approach that quantifies tumor phenotypic characteristics and heterogeneity by analyzing features extracted from medical images, holds promise as a potential biomarker for personalized cancer treatment and prognosis assessment (11-16). Researches focus on ONM diagnosis have achieve as high as 0.972 of area under the curve (AUC) by using radiomics-based diagnostic model (17). However, Zhang et al.’s research reported that the AUC of radiomics model in diagnosing ONM was only 0.813 (18). The inconsistence in diagnostic efficacy hinders the clinical application of radiomics models.
Machine learning algorithms, including Bayes, decision tree (DT), least absolute shrinkage and selection operator (LASSO), logistic regression (LR), random forest (RF), support vector machine (SVM), and extreme gradient boosting (XGB), have been employed for radiomics modeling and are crucial for establishing radiomics models (19-22). Machine learning algorithms can be broadly classified into linear and non-linear models, depending on how they handle different types of data. Linear models, such as LR, are better suited for data that is linearly separable or has a linear trend, while non-linear models like RF, SVM are better suited for data that has non-linear or irregular patterns. Choosing the right type of model for the data can improve the accuracy and performance of machine learning tasks. Zheng et al. (23) applied CT radiomics models based on different machine learning algorithms and compared the differential efficacy of different models, and found that the fusion model constructed by SVM algorithm showed superior differential ability in distinguishing benign and malignant parotid tumors. However, currently no research has studied which machine learning algorithm is more suitable for establishing ONM diagnostic models.
Therefore, the objective of this study was to identify the optimal machine learning algorithm for radiomics modeling and to compare the performances of the radiomics model, the clinical-radiological model and the combined model incorporate both radiomics features and clinical-radiological features in preoperatively predicting ONM in clinical stage IA lung adenocarcinoma patients. Evaluation metrics, including AUC, accuracy, precision, sensitivity, specificity, and F1 score, were specifically employed to measure and compare the performance of these models. The insights gained will provide valuable strategies for preoperative ONM prediction in patients with clinical stage IA lung adenocarcinoma, contributing to improved clinical decision-making and patient prognosis. We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1578/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This retrospective study was approved by the Institutional Review Board of the Cancer Hospital, Chinese Academy of Medical Sciences (Approval No. NCC2022C-693), which waived the requirement for informed consent.
Patients
We reviewed data from patients with surgically resected clinical stage IA lung adenocarcinomas that were treated between October 2005 and January 2017 in our center. The inclusion criteria were as follows: (I) patients who underwent surgery for lung adenocarcinoma; (II) LN dissection; and (III) contrast-enhanced HRCT performed <2 weeks before surgical resection. The exclusion criteria were (I) multiple lesions or evidence of metastasis; (II) tumor diameter >3 cm; (III) LN diameter in the hila or mediastina >1 cm on HRCT; and (IV) malignancy history in the last 5 years. In total, 732 patients met the criteria, of whom 129 were pathologically LN-positive and 603 were pathologically LN-negative. Propensity score-matching (PSM) was performed between the LN-positive and LN-negative groups and included age, sex, and smoking status. Patients were matched based on PSM using the nearest-neighbor method with a matching ratio of 1:1. Ultimately, this study included a total of 258 patients, with 182 patients (91 LN-positive and 91 LN-negative) assigned to the training set and the remaining 76 patients (38 LN-positive and 38 LN-negative) forming the test set. The random division followed a 7:3 ratio, ensuring a representative distribution across both sets.
Clinicopathological characteristics
For our analysis, clinical characteristics, including age, sex, and smoking status (non-smoker, smoker), were evaluated. Tumors were classified according to the International Association for the Study of Lung Cancer Grading System, which divides lung adenocarcinoma into three grades: grade 1, mainly lepidic components with <20% high-grade patterns (solid, micropapillary, and complex glandular components); grade 2, acinar or papillary components with <20% high-grade components; and grade 3, high-grade components with >20% high-grade patterns (Table S1) (24). Epidermal growth factor receptor (EGFR) mutations were detected in tumor tissue and plasma deoxyribonucleic acid (DNA) samples from 151 patients using an amplification-refractory mutation system or direct DNA sequencing.
CT image acquisition, interpretation, and feature extraction
We acquired HRCT images with 8-, 16-, or 64-spiral CT scanners (LightSpeed Ultra, ProSpeed, Discovery ST, or LightSpeed VCT; GE Medical Systems, Chicago, IL, USA). All patients underwent enhanced HRCT examination, in which 60–80 mL of intravenous contrast was administered at 2.0–2.5 mL/s and enhanced images were obtained 25–30 s after contrast infusion. HRCT images were obtained at 120 kVp and 250–350 mA with reconstruction using a standard algorithm. The reconstruction thickness was 0.625 or 1.25 mm, and the interval was 0.8–1.0 mm.
The radiological features of the primary tumor were assessed by two experienced thoracic radiologists (L.Z. and M.L.) both with over 10 years of experience in chest CT interpretation. These radiologists were blinded to all clinical and outcome information to ensure an unbiased evaluation. The evaluated features included tumor diameter, nodule consistency [pure ground glass nodule (pGGN), part solid nodule (PSN), solid nodule (SN)], bubble-like lucency, bronchiectasis, deep lobulation, emphysema, necrosis, pleural indentation, sharpness, lobar location, and tumor location [central (the inner third) or peripheral (the outer two-thirds of the lung fields)]. For the quantitative characteristics, the average values measured by two radiologists were used. Discrepancies in the interpretation of morphological features were resolved by a final consensus through group discussion. All measurements followed the recommendations of the Fleischer Society for CT-based measurement of pulmonary nodules (25).
Manual segmentation of the primary tumor was performed independently by a thoracic radiologist (L.Z.) and confirmed by another thoracic radiologist (M.L.). Discrepancies in tumor borders were resolved by a final consensus through group discussion. Tumor regions of interest were defined using open-source software (ITK-SNAP; http://www.itksnap.org) with lung window settings across all two-dimensional sections in the axial view. The window and level settings were varied to properly annotate nodule borders for nodules near the mediastinum or chest wall.
Radiomics features were extracted using Artificial Intelligence Kit software (A.K. V3.0.0. R, GE), which conformed to the image biomarker standardization initiative (26). First, we used linear interpolation to resample all images to a uniform voxel size of 1 mm × 1 mm × 1 mm to minimize the effects of different layer thicknesses. Second, based on the grayscale discretization process (bin width =25 for CT), we converted continuous images into discrete values. Finally, we used Gaussian Laplacian and wavelet image filters to eliminate mixed noise during image digitization to obtain low- or high-frequency features. The formula for calculating the radiomics signatures is available on the official documentation website (https://pyradiomics.readthedocs.io/en/latest/features.html). The minimum redundancy maximum relevance algorithm was initially applied to eliminate redundant and irrelevant features, resulting in a preliminary selection of 20 features. Subsequently, the LASSO method was used to further refine the subset of radiomics features, thus optimizing the selection process.
Model building and validation
Seven machine learning algorithms commonly used in radiomics (Bayes, LASSO, LR, DT, RF, SVM, and XGB) were used for radiomics feature selection and classification in the training cohort. Univariate LR analysis was conducted on the clinical and radiological features to identify significant features, with a significance level of P<0.1. These significant features were then used in multivariate LR analysis to obtain the ultimate predictor variables for the development of the clinical model. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. A combined model was built by integrating the optimal radiomics and clinical models. The development and validation of the model were performed by a biomedical engineering expert (Y.M.W.), who had no access to any clinical or outcome data to guarantee a fair assessment.
Statistical analyses
Frequency distribution and descriptive statistics were determined for all variables. Data were expressed as means ± standard deviations when normally distributed or as medians [interquartile ranges (IQRs)] when normality assumptions were not met. The Kolmogorov-Smirnov test was used to test the normality assumptions. The equivalence of patient attributes between the training and test cohorts was analyzed using the t-test or Wilcoxon rank-sum test for continuous variables and Chi-squared test or Fisher’s exact test for categorical variables. The diagnostic performance of the models was assessed using various metrics, including the AUC of the receiver operating characteristic (ROC) curve, accuracy, precision, sensitivity, specificity, and F1 score. Calibration plots were used to assess the alignment between predicted and observed values. The Brier score was used to evaluate the overall accuracy and calibration performance of the models. Decision curve analysis (DCA) was performed to evaluate the robustness and clinical applicability of the models. Statistical analyses were performed by Y.M.W. using IBM SPSS Statistics for Windows, version 25.0 (IBM Corp., Armonk, NY, USA) and R software (version 4.1.1; The R Foundation for Statistical Computing, Vienna, Austria). P<0.05 was considered statistically significant.
Results
Patient characteristics
This study included a total of 258 patients (Figure 1). The training cohort comprised 182 patients, whereas the test cohort included 76. The clinical and radiological features did not differ significantly between the training and test cohorts (all P>0.05) (Table S2). The median age of the patients was 59 (IQR, 51–66) years, 116 (45.0%) patients were male individuals, and 142 (55.0%) were female individuals. Most patients were non-smokers (n=173, 67.1%). The median tumor size was 1.9 (IQR, 1.5–2.5) cm. Most lesions on CT images were SNs (n=144, 55.8%), followed by PSNs (n=102, 39.5%), and pGGNs (n=12, 4.7%). Most tumors were classified as grade 2 (n=147, 57.0%), followed by grade 3 (n=74, 28.7%) and grade 1 (n=37, 14.3%). A median of 15 (IQR, 10–21) LNs was resected. Pathological stage N0 was observed in half (n=129, 50.0%) of the patients; 77 (29.8%) had N1 disease and 52 (20.2%) had N2 disease. The baseline characteristics are shown in Table 1 and Table S2.
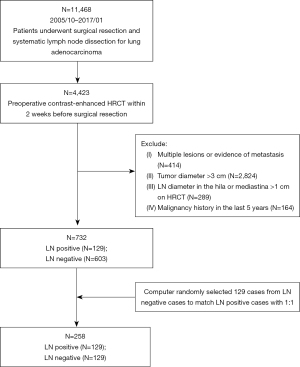
Table 1
| Characteristic | Values |
|---|---|
| Sex | |
| Male | 116 (45.0) |
| Female | 142 (55.0) |
| Age (years)† | 59 [51–66] |
| Smoking status | |
| Non-smoker | 173 (67.1) |
| Smoker | 85 (32.9) |
| Diameter (cm)† | 1.9 [1.5–2.5] |
| Nodule consistency | |
| pGGN | 12 (4.7) |
| PSN | 102 (39.5) |
| SN | 144 (55.8) |
| Boundaries | |
| Clear | 232 (89.9) |
| Fuzzy | 26 (10.1) |
| Bronchiectasis | |
| Absent | 251 (97.3) |
| Present | 7 (2.7) |
| Bubble-like lucency | |
| Absent | 163 (63.2) |
| Present | 95 (36.8) |
| Emphysema | |
| Absent | 217 (84.1) |
| Present | 41 (15.9) |
| Deep lobulation | |
| Absent | 218 (84.5) |
| Present | 40 (15.5) |
| Necrosis | |
| Absent | 199 (77.1) |
| Present | 59 (22.9) |
| Pleural retraction | |
| Absent | 131 (50.8) |
| Present | 127 (49.2) |
| Sharpness | |
| Round | 187 (72.5) |
| Irregular | 71 (27.5) |
| Invasive lobe | |
| Right upper | 89 (34.5) |
| Right middle | 20 (7.8) |
| Right lower | 46 (17.8) |
| Left upper | 67 (26.0) |
| Left lower | 36 (14.0) |
| Location | |
| Central | 70 (27.1) |
| Peripheral | 188 (72.9) |
| Surgical procedure | |
| Sublobectomy | 12 (4.7) |
| Lobectomy | 246 (95.3) |
| Adjuvant therapy | |
| Surgery alone | 164 (63.6) |
| Surgery plus adjuvant therapy | 94 (36.4) |
| Pathological T stage‡ | |
| T1is | 10 (3.9) |
| T1a | 31 (12.0) |
| T1b | 83 (32.2) |
| T1c | 58 (22.5) |
| T2a | 76 (29.5) |
| Pathological N stage‡ | |
| N0 | 129 (50.0) |
| N1 | 77 (29.8) |
| N2 | 52 (20.2) |
| Pathological stage‡ | |
| 0 (Tis) | 10 (3.9) |
| IA1 | 29 (11.2) |
| IA2 | 39 (15.1) |
| IA3 | 16 (6.2) |
| IB | 35 (13.6) |
| IIB | 77 (29.8) |
| IIIA | 52 (20.2) |
| Grading system of lung adenocarcinoma | |
| Grade 1 | 37 (14.3) |
| Grade 2 | 147 (57.0) |
| Grade 3 | 74 (28.7) |
| No. of resected lymph nodes† | 15 [10–21] |
| EGFR mutation | |
| Negative | 47 (31.1) |
| Positive | 104 (68.9) |
Unless otherwise indicated, data in parentheses are presented as percentages. †, data in brackets denote interquartile ranges; ‡, 8th staging classification. Overall, 151 patients underwent genetic testing. pGGN, pure ground glass nodule; PSN, part solid nodule; SN, solid nodule; EGFR, epidermal growth factor receptor.
Radiomics feature extraction and model construction
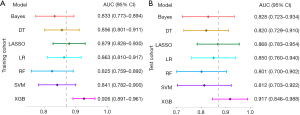
The study extracted 107 quantitative features from the imaging data, encompassing 18 first-order features, 16 gray-level run-length matrix features, 16 gray-level size zone matrix features, 24 gray-level co-occurrence matrix features, 14 shape (three-dimensional) features, 5 neighboring gray-tone difference matrix features, and 14 gray-level dependence matrix features. To determine their importance, the minimum redundancy maximum relevance algorithm was employed, resulting in the selection of the top 20 features for further analysis. Using the LASSO method, the features were further reduced to six (Table S3, Figure S1). Among the seven radiomics models evaluated, the XGB model exhibited the best performance, with an AUC of 0.926 (95% CI: 0.891–0.961) in the training cohort and 0.917 (95% CI: 0.846–0.988) in the test cohort (Figure 2A,2B).
Clinical-radiological factors associated with ONM
Univariate analysis of the training cohort revealed that diameter, nodule type, boundaries, bronchiectasis, bubble-like lucency, emphysema, deep lobulation, necrosis, pleural retraction, sharpness, and location were significantly associated with ONM (all P<0.1). In the multivariable LR analysis, nodule type [OR =0.390; 95% CI: (0.301, 0.478); P<0.001], boundaries [OR =0.202; 95% CI: (0.040, 0.363); P=0.015], necrosis [OR =0.234; 95% CI: (0.111, 0.356); P<0.001], sharpness [OR =0.156; 95% CI: (0.047, 0.264); P<0.001], and location [OR =−0.112; 95% CI: (−0.222, 0.001); P=0.048] were independent risk factors for ONM (Table 2). Based on these independent risk factors, a clinical model was developed that yielded an AUC of 0.855 (95% CI: 0.801–0.909) and 0.814 (95% CI: 0.718–0.911) in the training and test cohorts, respectively (Figure 3A,3B, Table 3).
Table 2
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| Sex | 1.000 (0.612, 1.633) | >0.999 | – | – | |
| Age | 1.005 (0.980, 1.030) | 0.719 | – | – | |
| Smoking status | 1.192 (0.709, 2.005) | 0.508 | – | – | |
| Diameter | 3.023 (1.948, 4.692) | <0.001 | – | – | |
| Nodule type | 12.030 (6.628, 21.836) | <0.001 | 0.390 (0.301, 0.478) | <0.001 | |
| Boundaries | 3.010 (1.219, 7.435) | 0.017 | 0.202 (0.040, 0.363) | 0.015 | |
| Bronchiectasis | 6.244 (0.741, 52.616) | 0.092 | – | – | |
| Bubble-like lucency | 1.770 (1.061, 2.955) | 0.029 | – | – | |
| Calcification | 3.048 (0.313, 29.691) | 0.337 | – | – | |
| Emphysema | 5.199 (2.296, 11.776) | <0.001 | – | – | |
| Deep lobulation | 4.218 (1.917, 9.281) | <0.001 | – | – | |
| Necrosis | 8.439 (3.929, 18.124) | <0.001 | 0.234 (0.111, 0.356) | <0.001 | |
| Pleural retraction | 1.929 (1.176, 3.164) | 0.009 | – | – | |
| Sharpness | 2.731 (1.537, 4.854) | 0.001 | 0.156 (0.047, 0.264) | <0.001 | |
| Location | 0.412 (0.233, 0.730) | 0.002 | −0.112 (−0.222, 0.001) | 0.048 | |
OR, odds ratio; CI, confidence interval.
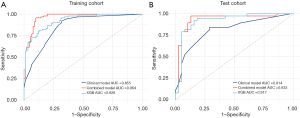
Table 3
| Model | AUC | Brier score | Accuracy | Precision | Sensitivity | Specificity | F1 score |
|---|---|---|---|---|---|---|---|
| XGB | |||||||
| Training | 0.926 (0.891–0.961) | 0.115 (0.066–0.164) | 0.841 | 0.860 | 0.813 | 0.868 | 0.836 |
| Test | 0.917 (0.846–0.988) | 0.116 (0.040–0.193) | 0.842 | 0.933 | 0.737 | 0.947 | 0.824 |
| Clinical | |||||||
| Training | 0.855 (0.801–0.909) | 0.150 (0.094–0.206) | 0.802 | 0.743 | 0.923 | 0.681 | 0.824 |
| Test | 0.814 (0.718–0.911) | 0.168 (0.076–0.260) | 0.776 | 0.744 | 0.842 | 0.711 | 0.790 |
| Combined | |||||||
| Training | 0.964 (0.938–0.989) | 0.078 (0.037–0.118) | 0.929 | 0.906 | 0.956 | 0.901 | 0.930 |
| Test | 0.933 (0.867–1.000) | 0.092 (0.024–0.160) | 0.868 | 0.889 | 0.842 | 0.895 | 0.865 |
Data in parentheses are confidence intervals. AUC, area under the curve; XGB, extreme gradient boosting.
Performance and robustness of the combined model
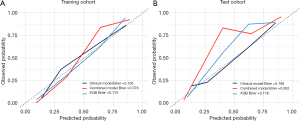
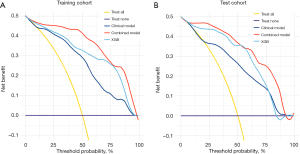
In the training cohort, the combined model showed exceptional performance, with an AUC of 0.964 (95% CI: 0.938–0.989) (Figure 3A, Table 3). Its robustness was further confirmed in the test cohort, which achieved a strong AUC of 0.933 (95% CI: 0.867–1.000) (Figure 3B, Table 3). The confusion matrix results highlighted the predictive accuracy of both the XGB and combined models for ONM, with the combined model demonstrating improved accuracy, sensitivity, and F1 scores in the test cohort (Table 3). Calibration curves showed the reliable performance of the three models, demonstrating a close match between the predicted ONM and actual ONM probabilities, as indicated by Brier scores of <0.25 (Figure 4A,4B, Table 3). The DCA revealed that the combined model provided a greater net benefit across a range of threshold probabilities, surpassing the performance of the XGB model (Figure 5A,5B).
Discussion
In this study, we compared seven machine learning algorithms for radiomics modeling and evaluated the performance of the optimal radiomics model and a combined model in predicting ONM in clinical stage IA lung adenocarcinoma. We found that XGB was the best algorithm for radiomics modeling and that the combined model had higher accuracy and robustness than the radiomics-only model.
In the 1990s, mediastinal LN dissection emerged as a potential strategy to improve the survival rates of patients with lung cancer, particularly when most patients had LNM (27). However, as the detection rates for early-stage lung cancer have significantly improved owing to advances in imaging technology, the necessity for LN dissection in patients with clinical stage IA lung adenocarcinoma is debated. This procedure carries risks such as damage to the blood and neurolymphatic vessels, leading to complications, including bleeding, nerve damage, and lymphedema. Allen et al. (28) found that up to 20% of patients with lung cancer undergoing LN dissection experience these complications, which considerably affect their quality of life and recovery time. Moreover, the presence of LNM is a crucial factor in determining the appropriate treatment approach for patients with lung cancer. Therefore, the accurate prediction of ONM in clinical stage IA lung adenocarcinoma is of significant importance.
Previous studies investigated the predictive capabilities of various machine learning algorithms, such as LR and RF, using radiomics features or a combination of clinical and radiological features to predict LNM (17,29). Our results align with these previous findings, as we also observed that the radiomics model could be used to predict LNM and that the combined model incorporating radiomics and clinical-radiological features outperformed the single-radiomics model. The combined model exhibited higher AUC, accuracy, sensitivity, and F1 score than the XGB model. Furthermore, DCA highlighted the clinical benefits of the combined model. These findings underscore the potential advantages of integrating multiple data sources into ONM predictive models, thereby offering improved accuracy and clinical utility.
What distinguishes our research is the extensive comparison of seven distinct machine-learning algorithms: Bayes, DT, LASSO, LR, SVM, RF, and XGB. Our results demonstrated the highest performance for XGB among these models, with an AUC of 0.917 in the test cohort. XGB is a scalable and efficient tree-boosting system that has been widely used in various domains, including web searches, recommendation systems, and bioinformatics (30). The XGB model has several advantages over other machine learning models, such as its ability to handle missing values, to prevent overfitting, to support parallel computing, and to provide feature importance scores. In contrast to commonly used models in radiomics modeling, such as LR and RF, the XGB model excels in capturing intricate and nonlinear relationships between features and outcomes through its utilization of gradient-boosting algorithms and regularization techniques. However, it is important to consider the outstanding performance of XGB within the specific context of our dataset and study population, as the results may vary when applied to different datasets and clinical scenarios. Nevertheless, our findings underscore the immense potential of the integrated XGB algorithm for accurately predicting ONM in clinical stage IA lung adenocarcinomas.
The study limitations included the relatively small sample size due to the low ONM rate in clinical stage IA lung adenocarcinoma. Multicenter studies may be adopted for validation in clinical applications. Another limitation was that we did not demonstrate the associations between biological processes and radiomics in the ONM process, which requires further research.
Conclusions
Our study results showed that XGB was superior to other machine learning algorithms in predicting ONM in clinical stage lung adenocarcinoma in radiomics modeling and that incorporating radiomics and clinical-radiological features helped improve the model’s performance.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1578/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1578/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1578/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1578/coif). Y.M.W. is an employee of GE HealthCare China, Pudong New Town, Shanghai, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of board of the Cancer Hospital, Chinese Academy of Medical Sciences (No. NCC2022C-693) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Sateia HF, Choi Y, Stewart RW, et al. Screening for lung cancer. Semin Oncol 2017;44:74-82. [Crossref] [PubMed]
- Li F, Zhai S, Fu L, et al. Nomograms for intraoperative prediction of lymph node metastasis in clinical stage IA lung adenocarcinoma. Cancer Med 2023;12:14360-74. [Crossref] [PubMed]
- Deng J, Zhong Y, Wang T, et al. Lung cancer with PET/CT-defined occult nodal metastasis yields favourable prognosis and benefits from adjuvant therapy: a multicentre study. Eur J Nucl Med Mol Imaging 2022;49:2414-24. [Crossref] [PubMed]
- Huang X, Wang J, Chen Q, et al. Mediastinal lymph node dissection versus mediastinal lymph node sampling for early stage non-small cell lung cancer: a systematic review and meta-analysis. PLoS One 2014;9:e109979. [Crossref] [PubMed]
- Shayani J, Flores RM, Hakami A. Mediastinal lymph node dissection: the debate is not resolved. J Thorac Dis 2017;9:1848-50. [Crossref] [PubMed]
- Darling GE, Allen MS, Decker PA, et al. Randomized trial of mediastinal lymph node sampling versus complete lymphadenectomy during pulmonary resection in the patient with N0 or N1 (less than hilar) non-small cell carcinoma: results of the American College of Surgery Oncology Group Z0030 Trial. J Thorac Cardiovasc Surg 2011;141:662-70. [Crossref] [PubMed]
- Gao Z, Wang X, Zuo T, et al. A predictive nomogram for lymph node metastasis in part-solid invasive lung adenocarcinoma: A complement to the IASLC novel grading system. Front Oncol 2022;12:916889. [Crossref] [PubMed]
- Li W, Zhou F, Wan Z, et al. Clinicopathologic features and lymph node metastatic characteristics in patients with adenocarcinoma manifesting as part-solid nodule exceeding 3 cm in diameter. Lung Cancer 2019;136:37-44. [Crossref] [PubMed]
- Wang Z, Wu Y, Wang L, et al. Predicting occult lymph node metastasis by nomogram in patients with lung adenocarcinoma ≤2 cm. Future Oncol 2021;17:2005-13. [Crossref] [PubMed]
- Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012;264:387-96. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Larue RT, Defraene G, De Ruysscher D, et al. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 2017;90:20160665. [Crossref] [PubMed]
- Zhang L, Lv L, Li L, et al. Radiomics Signature to Predict Prognosis in Early-Stage Lung Adenocarcinoma (≤3 cm) Patients with No Lymph Node Metastasis. Diagnostics (Basel) 2022;12:1907. [Crossref] [PubMed]
- Zhong Y, Yuan M, Zhang T, et al. Radiomics Approach to Prediction of Occult Mediastinal Lymph Node Metastasis of Lung Adenocarcinoma. AJR Am J Roentgenol 2018;211:109-13. [Crossref] [PubMed]
- Zhang R, Zhang R, Luan T, et al. A Radiomics Nomogram for Preoperative Prediction of Clinical Occult Lymph Node Metastasis in cT1-2N0M0 Solid Lung Adenocarcinoma. Cancer Manag Res 2021;13:8157-67. [Crossref] [PubMed]
- Dong F, Li Q, Xu D, et al. Differentiation between pilocytic astrocytoma and glioblastoma: a decision tree model using contrast-enhanced magnetic resonance imaging-derived quantitative radiomic features. Eur Radiol 2019;29:3968-75. [Crossref] [PubMed]
- Li J, Li X, Ma J, et al. Computed tomography-based radiomics machine learning classifiers to differentiate type I and type II epithelial ovarian cancers. Eur Radiol 2023;33:5193-204. [Crossref] [PubMed]
- Lin Z, Wang T, Li Q, et al. Development and validation of MRI-based radiomics model to predict recurrence risk in patients with endometrial cancer: a multicenter study. Eur Radiol 2023;33:5814-24. [Crossref] [PubMed]
- Xu J, Guo J, Yang HQ, et al. Preoperative contrast-enhanced CT-based radiomics nomogram for differentiating benign and malignant primary retroperitoneal tumors. Eur Radiol 2023;33:6781-93. [Crossref] [PubMed]
- Zheng Y, Zhou D, Liu H, et al. CT-based radiomics analysis of different machine learning models for differentiating benign and malignant parotid tumors. Eur Radiol 2022;32:6953-64. [Crossref] [PubMed]
- Moreira AL, Ocampo PSS, Xia Y, et al. A Grading System for Invasive Pulmonary Adenocarcinoma: A Proposal From the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 2020;15:1599-610. [Crossref] [PubMed]
- Bankier AA, MacMahon H, Goo JM, et al. Recommendations for Measuring Pulmonary Nodules at CT: A Statement from the Fleischner Society. Radiology 2017;285:584-600. [Crossref] [PubMed]
- Zwanenburg A, Vallières M, Abdalah MA, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295:328-38. [Crossref] [PubMed]
- Keller SM, Adak S, Wagner H, et al. Mediastinal lymph node dissection improves survival in patients with stages II and IIIa non-small cell lung cancer. Eastern Cooperative Oncology Group. Ann Thorac Surg 2000;70:358-65; discussion 365-6. [Crossref] [PubMed]
- Allen MS, Darling GE, Pechet TT, et al. Morbidity and mortality of major pulmonary resections in patients with early-stage lung cancer: initial results of the randomized, prospective ACOSOG Z0030 trial. Ann Thorac Surg 2006;81:1013-9; discussion 1019-20. [Crossref] [PubMed]
- Cong M, Feng H, Ren JL, et al. Development of a predictive radiomics model for lymph node metastases in pre-surgical CT-based stage IA non-small cell lung cancer. Lung Cancer 2020;139:73-9. [Crossref] [PubMed]
- Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. KDD '16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM, 2016.


