
ALAS2 overexpression alleviates oxidative stress-induced ferroptosis in aortic aneurysms via GATA1 activation
Highlight box
Key findings
• 5-aminolevulinate synthase 2 (ALAS2) and GATA binding protein 1 (GATA1) play critical roles in the pathophysiology of aortic aneurysms.
• Overexpression of ALAS2 reduces oxidative stress and ferroptosis in aortic aneurysm cells.
• GATA1 knockdown partially reversed the protective effect of ALAS2 on ferroptosis.
What is known and what is new?
• Aortic aneurysms have a complex etiology, with increasing incidence and significant mortality. Ferroptosis is implicated in various clinical diseases, including aortic aneurysms.
• This study is novel in demonstrating the interaction between ALAS2 overexpression and GATA1 knockdown in the context of ferroptosis in aortic aneurysms.
What is the implication, and what should change now?
• ALAS2 holds potential as a diagnostic marker and therapeutic target for aortic aneurysms.
• Understanding the interaction between ALAS2 and GATA1 could guide new therapeutic strategies that may improve patient outcomes in aortic aneurysm treatment.
Introduction
The bulging of an aorta caused by the compression, traction, or erosion of surrounding tissues by a tumor is known as an aortic aneurysm (1). It may appear anywhere on the aorta and may be tube-shaped or bulb-shaped (2). It is generally defined as a focal aortic diameter exceeding 50% of the expected normal diameter (3). Clinically, aortic aneurysms are mainly divided into abdominal aortic aneurysms, which occur in the abdominal part of the aorta, and thoracic aortic aneurysms (TAA), which are located in the chest cavity (4,5). Several factors contribute to the pathogenesis of aortic aneurysms, including genetic predisposition, hypertension, atherosclerosis, and connective tissue disease (6,7).
The exact prevalence and death toll of aortic aneurysms varies by region and population; however, extensive research has shown that the incidence of aortic aneurysms is increasing (5). Thus, early detection and intervention are critical to reduce the risk of aortic rupture, dissection, and other life-threatening complications (8). Current treatment strategies primarily include surgical repair and medical management (mostly by blood pressure lowering drugs to mitigate) to mitigate the risk of aneurysm expansion and rupture (9,10). Despite advancements, there remains a pressing demand to identify novel diagnostic biomarkers, therapeutic strategies such as gene therapy and stem cell treatments, as well as prognostic indicators to enhance the precision and efficacy of aortic aneurysm interventions. This necessitates concerted research efforts in emerging fields like gene therapy, stem cell therapy, and novel drug development.
A new type of cell death called ferroptosis is critical in many clinical diseases (11,12). In the context of aortic aneurysms, research into the involvement, mechanisms, and therapeutic potential of ferroptosis has gained prominence. Using bioinformatics technology, Ren et al. identified key ferroptosis-related genes related to abdominal aortic aneurysm formation and rupture, including glutathione peroxidase 4 (GPX4), solute carrier family 2 member 1 (SLC2A1), and phosphatidylethanolamine binding protein 1 (PEBP1) (13). Further, research by Filiberto et al. demonstrated that the immunomodulatory effects of Resolvin D1 are mediated through macrophage formyl peptide receptor 2. This regulation effectively reduced ferroptosis and high-mobility group box 1 release, thereby attenuating aortic inflammation and remodeling in the pathogenesis of abdominal aortic aneurysms (14). Loick et al. found that ferritin is elevated in abdominal aortic aneurysms (AAA) and negatively correlates with lipid-cotransport protein-2 (LCN2). Smooth muscle cell (SMC)-specific ferritin-deficient mice display a more severe AAA phenotype. LCN2-neutralising antibodies restore autophagy, reduce neutrophil infiltration, and prevent the elevation of the AAA phenotype, attenuating the progression of the pathology (15).
5-aminolevulinate synthase 2 (ALAS2) encodes an enzyme responsible for the initial and rate-limiting steps of heme biosynthesis (16). Where heme is a component of the iron within the hemoglobin molecule (17). This suggested that the activity and regulation of ALAS2 is critical for heme synthesis and intracellular iron utilization. The study by Sawicki et al. identified increased oxidative stress and cell death associated with ALAS2 overexpression due to heme accumulation. They further observed that knockdown of ALAS2 reversed the increase in heme content and cell death in cultured cardiomyoblasts exposed to hypoxic conditions. By administering the mitochondrial antioxidant MitoTempo to ALAS2 overexpressing cardiomyoblasts, elevated levels of oxidative stress and cell death could be normalised to baseline, suggesting that the effects of ALAS2 and increased haemoglobin are mediated through elevated mitochondrial oxidative stress (18). A study by Pilling et al. revealed an age- and sex-specific gene expression signature for muscle strength that may be present in the blood. They found that gene expression levels correlated with strength after adjustment for cofactors and several statistical tests, including ALAS2 (19). Peng et al. showed that ALAS2 overexpressing transgenic mice (Tg mice) exhibited muscular dystrophy, which was associated with reductions in atrogin-1 and MuRF-1, as well as a strong association with mitochondrial dysfunction (20). The study by Reinwald et al. identified 222 genes with early responses to specific major modes of action (MoA) in environmental non-target organisms. Many of these genes are associated with three major processes: cardiomyocyte development and function, oxygen transport and hypoxic stress, and neuronal development and plasticity, including ALAS2, which is associated with oxygen transport and hypoxic stress (21). A study by Massaiu et al. found higher expression levels of heme synthesis and externalisation (ALAS2 upregulation) in left ventricular samples from subjects with dilated cardiomyopathy (DCM), which was confirmed by bulk and single nucleus RNA sequencing (RNA-seq) data (22). The findings provided above suggest that ALAS2 plays an important role in several biological processes, including oxidative stress, muscle nutrition, oxygen transport and hypoxic stress, which provides important clues to further understand its role in the development and treatment of related diseases. Currently, the interaction between ALAS2 and ferroptosis is an emerging and important research area, but its mechanism of action in aortic aneurysms is still unclear. Therefore, the relationship between them needs to be explored to improve our understanding of aortic aneurysms and reveal potential new treatment strategies.
By integrating bioinformatics analysis with in vitro research, we sought to gain an understanding of the function of ferroptosis in the etiology of aortic aneurysms. Specifically, the interaction between ALAS2 and the transcription factor GATA binding protein 1 (GATA1) and their effects on ferroptosis and apoptosis of mouse aortic vascular smooth muscle cells (MOVAS) induced by hydrogen peroxide (H2O2) were examined. Our findings highlighted the importance of ALAS2 as a potential diagnostic marker and therapeutic target for aortic aneurysms, which could possibly enhance diagnosis and therapy. We present this article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-370/rc).
Methods
Downloading and processing of the GSE9106 dataset
The R program (version 4.0.1) was used to preprocess a microarray dataset for TAAs (GSE9106) that was retrieved from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/gds/). The dataset includes TAA samples (n=59) and their corresponding controls (n=34). Using the GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/), the probe identifier (probe ID) was converted to the gene symbol and a differential analysis was carried out. The threshold standard for fold change (FC) was >1.5 or <0.67, and the modified P value was <0.05. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Weighted gene co-expression network analysis (WGCNA)
A WGCNA was conducted to analyze the differentially expressed genes (DEGs) in the GSE9106 data set. A gene co-expression network was created using the R language’s “WGCNA” package (version 3.5.1). The soft threshold power was precisely tuned at β =12 to guarantee a scale-free topology. A topological overlap matrix (TOM) was generated from the weighted adjacency matrix, which served as a robust measure of network connectedness after the network had been built. Hierarchical clustering is performed based on TOM to generate gene modules, which are visualized through dendrograms. In this structure, individual branches (shown in different colors) represent different gene modules. Using weighted correlation coefficients, DEGs exhibiting similar expression trajectories were merged into corresponding modules. Ultimately, we examined the association between the gene modules and different samples of GSE9106 to determine the key module.
Analysis of protein-protein interactions (PPIs)
To elucidate the potential PPIs in the turquoise module, we conducted a PPI network analysis using the Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) database, along with Cytoscape software (Version 3.8.1). The generated PPI network, comprising the Molecular Complex Detection (MOCODE), Maximal Clique Centrality (MCC), and Maximal Neighborhood Component (MNC) networks, was visualized using the open-source network visualization software platform, Cytoscape. This visualization facilitated a detailed analysis of the PPIs within the module.
Identification and expression analysis of intersecting genes
Bioinformatics and Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to identify the intersecting genes in the MOCODE, MCC, and MNC networks. After identifying the intersecting genes, the expression levels of these intersecting genes in the TAA and control samples of the GSE9106 dataset were detected. A P value <0.05 was considered statistically significant.
Expression of GATA1 and correlation analysis with ALAS2
To explore the function of GATA1 in TAAs, GATA1 expression in the control and TAA samples in the GSE9106 dataset was first examined. Next, a correlation analysis was performed to examine the expressions of GATA1 and ALAS2 in the Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) database, and the Pearson correlation coefficients were calculated to evaluate the strength and direction of the linear relationships between the genes. A P value <0.05 was considered statistically significant.
Just Another Spar Promoter Analysis Resource (JASPAR) database
The JASPAR database (http://jaspar.genereg.net/) comprehensively collects transcription factor binding profiles and matrices. It provides valuable insights into potential binding motifs that transcription factors may recognize in gene promoter regions. In this study, the JASPAR database was used to predict the ALAS2 and GATA1 binding locations.
Cell lines and cell culture
The MOVAS were provided by the American Type Culture Collection (ATCC; Manassas, VA, USA). The MOVAS were maintained in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum (FBS) to which 1% penicillin-streptomycin was added. The cells were then cultivated in a humid environment at 37 °C with 5% carbon dioxide.
Transfection assay
H2O2 (800 µmol/L) was used to induce the damage model of the MOVAS. For the purpose of transient transfection, the MOVAS were inoculated into 24-well plates at an adjusted density of 2×105 cells per well. A plasmid encoding ALAS2 was transfected into the MOVAS using an appropriate transfection method to enable the transfected cells to express the ALAS2 protein for a specific period to achieve overexpression. Next, to effectively knock down the expression of GATA1, a particular small interfering RNA targeting GATA1 was transfected into the MOVAS. The cells were then incubated for the required amount of time. Lipofectamine 3000 (Invitrogen, MA, USA) was used to transfect the cells in accordance with the manufacturer’s instructions.
Quantitative real-time-polymerase chain reaction (qRT-PCR) assay
The total RNA of the MOVAS was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. A PrimeScript RT kit (Takara, Tokyo, Japan) was used for the complementary DNA synthesis. SYBR Green PCR Master Mix (Applied Biosystems, CA, USA) was used to perform the qRT-PCR with the StepOnePlus Real-Time PCR System (Applied Biosystems). The 2−ΔΔCT technique was used to examine the data, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) abundance was used as a standard. The following primer sequences were used in the amplification process: ALAS2 forward: 5'-CTGCCAGGGTGCGAGATT-3', reverse: 5'-TTGGCTGCTCCACTGTTACG-3'. GATA1 forward: 5'-CCTGCTTTGTTGCCAATG-3', reverse: 5'-CTGCTCCACTGTTACGGATAC-3'. The primer sequences used for amplifying GAPDH were: GAPDH forward: 5'-CAAGCTCATTTCCTGGTATGAC-3', reverse: 5'-CAGTGAGGGTCTCTCCTTCCT-3'.
Westtern blotting (WB) assay
Radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific, IL, USA) was used to make the cellular protein lysates, which also included protease and phosphatase inhibitors (1:1,000) from the same supplier. The protein concentration (2 mg/mL) was quantified using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). The same amounts of protein were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then put onto a polyvinylidene difluoride (PVDF) membrane (Millipore, MA, USA). Specific primary antibodies against ALAS2, NFE2 like BZIP transcription factor 2 (NRF2), solute carrier family 7 member 11 (SLC7A11), glutathione peroxidase 4 (GPX4), and GATA1 (Abcam, Cambridge, UK; 1:1,000) were used. As an internal reference, GAPDH (Abcam; 1:5,000) was employed. Following this, the membrane underwent an incubation phase with secondary antibodies linked to horseradish peroxidase (provided by Amersham Bioscience, Tokyo, Japan). Next, an advanced chemiluminescence kit was used to visualize the protein bands (Thermo Fisher Scientific, Waltham, MA, USA). A densitometric analysis was executed using a ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA). The intensity was measured using ImageJ software.
Cell counting kit-8 (CCK-8) assay
The viability of the cells was assessed using the CCK-8, (Dojindo, Kumamoto, Japan). The RPE cells were seeded at a density of 5×103 cells per well in 96-well plates. Following the treatment, each well received CCK-8 reagent. Subsequently, absorbance at 450 nm was measured at 24, 48, 72, and 96 hours using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Flow cytometry
For the flow cytometric analysis, the MOVAS were detached using trypsin- ethylenediaminetetraacetic acid (EDTA) (Gibco, Waltham, MA, USA) and washed with phosphate-buffered saline. In accordance with the manufacturer’s instructions, the cells were stained with Annexin V and propidium iodide to distinguish among the live, apoptotic, and necrotic cells. A flow cytometer (BD Biosciences, CA, USA) was used for the flow cytometry, and the data were analyzed using FlowJo software (FlowJo LLC, Oregon, USA) to quantify the cell apoptosis rate.
Ferrous iron assay
A 24-well plate was used to seed the MOVAS (2×103 cells/well). Intracellular ferrous iron levels were determined in accordance with the manufacturer’s instructions and using an iron colorimetric assay kit (Abcam).
Enzyme-linked immunosorbent assay (ELISA)
Appropriately diluted samples of the cell culture supernatant were added to the wells of an ELISA plate pre-coated with interleukin (IL)-1β, tumor necrosis factor α (TNF-α), and IL-6 antibodies. After incubation and washing, the enzyme-linked secondary antibody was added, followed by a chromogenic substrate. The reaction was stopped and the absorbance was measured at the appropriate wavelength using a microplate reader. The concentrations of inflammatory factors were determined by comparing the absorbance values to a standard curve generated using corresponding standards of known concentration.
Assessment of malonaldehyde (MDA), reactive oxygen species (ROS), and glutathione (GSH) levels
Following the H2O2 treatment and ALAS2 overexpression, the levels of the ROS, MDA, and GSH in the MOVAS were measured. A detection kit based on fluorescence was used to quantify the ROS. A thiobarbituric acid reactive substances test was used to evaluate the MDA levels. A colorimetric assay involving the reaction with 5,5'-Dithiobis (2-nitrobenzoic acid) (DTNB) was used to quantify GSH. Measurements were made using a calibrated microplate reader, and all processes were carried out in accordance with the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assay
A ChIP analysis was conducted to observe the association between proteins and DNA. After the DNA and proteins were cross-linked with formaldehyde, the cells were lysed. Chromatin was fragmented using sonication, and the cells were then incubated with primary antibodies against ALAS2, GATA1, and NC. The cross-links were reversed, DNA was extracted, and a quantitative PCR analysis was performed to identify the binding sites.
Dual-luciferase reporter assay
The cells were allocated into a 96-well format and subsequently transfected with pmirGLO vectors containing the sequences for ALAS2 3' untranslated region wild-type (ALAS2-3'UTR-WT) or ALAS2 3' untranslated region mutant (ALAS2-3'UTR-MT), in conjunction with small interfering RNA targeting GATA1 (si-GATA1). Post 48-hour transfection, luciferase expression was quantified using a dedicated assay kit, with the results normalized to the Renilla luciferase signal to ensure consistency. This procedure was replicated in triplicate to ensure the robustness of the findings.
Statistical analysis
SAS version 9.2 (SAS Institute Inc.) was used to examine the data. Each experiment was carried out in triplicate at least. A two-tailed Student’s t-test was used to evaluate differential expression and correlation coefficients. A P value <0.05 was considered statistically significant. All the tests were conducted in duplicate and were two-sided.
Results
Comprehensive analysis of DEGs and module-based network relationships in aortic aneurysms
In this study, 567 upregulated and 200 downregulated DEGs were identified from the control and TAA samples of the GSE9106 data set (Figure 1A). To determine the optimal soft threshold power for fitting the scale-free topology model, a no-scale fit analysis was conducted, resulting in a power of 12 (Figure 1B). Subsequently, we performed a comprehensive clustering analysis of 93 samples from the GSE9106 data set. The genes were clustered into different modules based on their co-expression patterns using the WGCNA method. Each module was represented by a specific color (Figure 1C). To assess the relationships between the identified modules, we explored the adjacency of hub genes. Notably, the turquoise module exhibited a significant correlation with sample characteristics, with a correlation coefficient of 0.41 (Figure 1D). PPI networks were subsequently constructed. The MCODE network contained 13 nodes and 51 edges (Figure 1E), while the MCC network featured 10 nodes and 44 edges (Figure 1F), and the MNC network included 10 nodes and 20 edges (Figure 1G).

Identification and characterization of intersecting genes
Three intersecting genes (i.e., ALAS2, GYPA, and GYPB) were identified from the MCODE, MCC, and MNC networks (Figure 2A). The subsequent individual expression analysis of these three genes revealed that they were significantly upregulated in the TAA group of the GSE9106 data set (Figures 2B-2D). Thus, these three genes may be potential aortic aneurysm genes. Based on bioinformatics analysis and literature review, ALAS2 was chosen for further investigation.

Iron content in MOVAS cells increases after H2O2 induction
Mitochondrial damage is a prominent feature of iron-dependent cell death. H2O2 (800 µmol/L) was employed to induce damage in the MOVAS. The MOVAS were treated with ferroptosis inhibitors [simvastatin (SIM) and ferrostatin-1 (Fer-1)] and catalysts [erastin and iron-saturated holo-transferrin (HTF)] to further examine the function of oxidative stress in the development of ferroptosis. H2O2 and Erastin significantly increased the iron levels, and the effect was more pronounced in cells treated with H2O2 + HTF. Conversely, Fer-1 and SIM alleviated the H2O2-induced increases in iron to a certain extent (Figure 3A). As Figure 3B shows, H2O2 reduced the GSH levels in comparison to the control group, and Fer-1 further exacerbated this result. The ferroptosis catalyst reversed the reducing effect of H2O2 on GSH levels to a certain extent. The results in Figure 3C indicated that H2O2 or erastin treatment resulted in increased ROS levels. After H2O2 treatment, the addition of Fer-1 or SIM resulted in a decrease in ROS levels, whereas the addition of HTF resulted in an increase in ROS content. Additionally, in the WB experiment, following the addition of H2O2, the level of ALAS2 increased, while the levels of ferroptosis negative regulators showed the opposite result. HTF promoted the H2O2-induced reduction of NRF2, SLC7A11, GPX4, and FTH1, while Fer-1 and SIM reversed this effect (Figure 3D,3E). These findings suggested that their effect in the modulation of ferroptosis warrants further investigation.
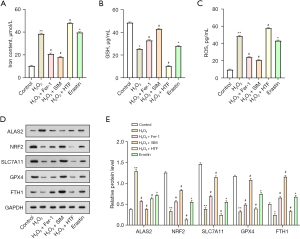
ALAS2 overexpression reverses the H2O2-induced inflammatory response and oxidative stress in MOVAS
The qRT-PCR and WB analysis results revealed a significant increase in ALAS2 expression levels after H2O2 treatment. Further, the overexpression of ALAS2 led to a more pronounced increase in ALAS2 expression (Figure 4A,4B). The ELISA results showed that the IL-1β, IL-6, and TNF-α levels in the H2O2-induced MOVAS were significantly upregulated. The overexpression of ALAS2 reversed these effects, resulting in lower levels of these inflammatory cytokines (Figure 4C-4E). The trends in the ROS and MDA changes corresponded with the alterations in the expression of inflammatory factors, showing a consistent pattern (Figure 4F,4G). Further, the pattern of GSH levels was contrary to that of ROS and MDA; the GSH levels decreased after adding H2O2, but when H2O2 acted synergistically with ALAS2 overexpression, the GSH levels increased (Figure 4H).
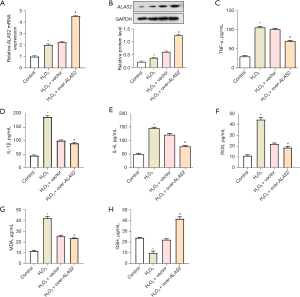
ALAS2 overexpression regulates apoptosis and iron concentration in H2O2-treated cells
The flow cytometry analysis showed that the H2O2 treatment significantly increased the apoptosis rate. However, the interaction with the overexpressed ALAS2 resulted in reduced apoptosis (Figure 5A,5B). Further, H2O2 treatment significantly increased cellular iron concentration, which subsequently decreased following the overexpression of ALAS2 (Figure 5C). Further, the H2O2 treatment resulted in a significant decrease in the protein levels of the ferroptosis negative regulators. The overexpression of ALAS2 partially restored the protein levels of these factors (Figure 5D,5E). All of these results suggested that the overexpression of ALAS2 may act as a protective mechanism against H2O2-induced ferroptosis by regulating the key proteins involved in ferroptosis.

GATA1 transcriptional activation ALAS2
The expression analysis of the GSE9106 dataset showed that GATA1 was significantly overexpressed in the TAA samples compared with the controls (Figure 6A). The GEPIA database revealed a positive correlation between this gene and ALAS2 expression (Figure 6B). The qRT-PCR and WB results showed that the GATA1 expression levels were significantly increased in the MOVAS after H2O2 treatment. Conversely, the knockdown of GATA1 resulted in a significant reduction in GATA1 expression (Figure 6C,6D). As Figure 6E shows, the binding site between GATA1 and ALAS2 was predicted using the JASPAR database (size of promoter fragment about 3,500 bp upstream). This prediction was confirmed by luciferase activity assays, which confirmed their interaction (Figure 6F). Further, GATA1 exhibited a strong correlation with the ALAS2 promoter region (Figure 6G), which suggested that GATA1 exerted a regulatory effect on ALAS2 expression under oxidative stress conditions.

Effects of ALAS2 and GATA1 on H2O2-induced inflammation and oxidative stress in MOVAS
The CCK-8 test revealed a considerable decrease in cell proliferation following H2O2 therapy. The overexpression of ALAS2 alleviated the decrease in cell proliferation induced by H2O2. However, the knockdown of GATA1 reversed the effect of ALAS2 overexpression and continued to cause a decrease in proliferation activity (Figure 7A). The ELISA results showed that the H2O2 treatment significantly upregulated the expression of inflammatory cytokines in the MOVAS. The combined treatment of H2O2, overexpressed ALAS2, and si-GATA1 further amplified the expression of these inflammatory markers compared with the expression of overexpressed ALAS2 alone (Figure 7B-7D). The levels of ROS and MDA, increased after H2O2 treatment and further increased in the presence of ALAS2 overexpression and GATA1 knockdown (Figure 7E,7F). Conversely, the levels of GSH showed the opposite trend, decreasing after H2O2 treatment and further decreasing with the knockdown of GATA1 in the presence of overexpressed ALAS2 (Figure 7G). These findings suggested that ALAS2 and GATA1 interact closely to control oxidative stress and inflammatory reactions in MOVAS.
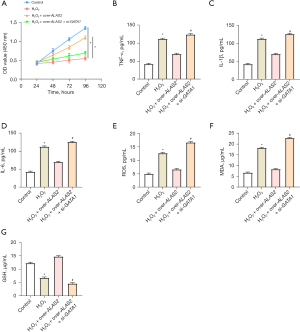
ALAS2 regulates iron content and ferroptosis in H2O2-treated MOVAS by activating GATA1
According to the iron colorimetric assay results, the MOVAS treated with H2O2 had a much lower iron content after ALAS2 overexpression and GATA1 knockdown than after H2O2 treatment alone. Conversely, the iron levels remained increased in comparison to the controls (Figure 8A). Under the same experimental circumstances, the WB analysis showed that H2O2 therapy downregulated the expression of the negative regulators of ferroptosis. Interestingly, after being treated with H2O2, the cells that had both ALAS2 overexpression and concomitant GATA1 knockdown showed a partial reversal in the expression levels of these ferroptosis-related proteins (Figure 8B,8C). This finding suggested that GATA1 and ALAS2 are important players in the cellular response to oxidative stress, especially in the regulation of iron levels and the alteration of the expression of important proteins that regulate ferroptosis.

Discussion
An aortic aneurysm is life-threatening due to the pathological dilation of the thoracic aorta, which, if left untreated, can lead to dissections or ruptures with high mortality rates (23,24). Traditional diagnostic techniques such as computed tomography and magnetic resonance imaging are critical to the treatment of clinical patient (25,26); however, they have limitations, including radiation exposure, high costs, and the use of contrast agents. As a result, there is a rising interest in discovering biomarkers, such as genetic markers, proteins, or other molecular indicators for the early, non-invasive detection of aortic aneurysms (27,28).
Advances in our understanding of the genetic underpinnings of aortic aneurysms have paved the way for targeted gene treatments. These therapies aim to address the root causes of aortic aneurysms by modulating key genes involved in aortic aneurysm development, offering the potential for more personalized and effective treatments. Presently, the 5-year survival rate for aortic aneurysm patients depends on factors such as the aneurysm size, location, and timely interventions (29). Surgical and endovascular procedures have shown efficacy in preventing aortic rupture, significantly improving survival rates (30,31). However, innovative therapeutic strategies, including gene-based treatments, may further enhance these rates. In summary, the identification of efficient and precise biomarkers for the diagnosis, treatment, and prognosis of aortic aneurysms is crucial.
In the present study, ALAS2 was identified as one of the key genes with significantly increased expression in the TAA samples, which suggests that this gene could be involved in the etiology of aortic aneurysms. Ferroptosis has emerged as a key factor in the progression of various diseases, highlighting the importance of iron regulation in cellular health and the potential to target ferroptosis in therapeutic strategies (32). The dysregulation of ALAS2 expression and its association with ferroptosis has been demonstrated in other diseases. ALAS2 activity is controlled by intracellular iron levels, which underscores the significance of maintaining adequate iron supplies for hemoglobin synthesis. For example, Ono et al. showed that ALAS2 mutations are associated with the occurrence of ferroptosis in X-linked sideroblastic anemia (33). Using a zebrafish model, Lv et al. (34) showed that insufficient hemoglobin production due to the abnormal function of ALAS2 induced ferroptosis in hematopoietic stem and progenitor cells. Whatley et al. (35) showed that C-terminal deletion of ALAS2 may result in gain-of-function and X-linked dominant protoporphyria without iron excess or anemia. Further, another study reported a link between ALAS2 and ferroptosis. Specifically, Song et al. found that ALAS2 expression was regulated by the ferroptosis response in a rat germinal matrix hemorrhage model, which may affect the molecular mechanisms of brain injury (36). These studies highlight the potential of ALAS2 to serve as an interventional target for a range of iron-related diseases.
Ferroptosis is a mode of controlled cell death closely related to oxidative stress (37). In this complex interaction, MDA functions as a crucial indicator of lipid peroxidation, which indicates cellular damage, and ROS initiate ferroptosis by causing damage to cellular components (38). Conversely, GSH serves as an essential antioxidant, and ferroptosis is characterized by its deficiency (39). Recent research by Zhou et al. showed that cordycepin targets vascular endothelial growth factor to inhibit apoptosis, inflammation, and oxidative stress in TAAs (40). Branchetti et al. discovered a potential link between increased peak wall stress, oxidative stress (ROS accumulation), and a shift in the vascular SMC phenotype towards a synthetic state, indicating the likely involvement of oxidative stress in TAA development (41). Further, Portelli et al. explored the association between oxidative stress and genetically triggered TAA, providing extensive evidence of oxidative stress and ROS imbalance (42).
The ferroptosis negative regulators NRF2, FTH1, GPX4, and SLC7A11 are critical for preserving iron metabolism and cellular redox homeostasis (43). Among them, NRF2 stimulates improved GSH production, facilitates ROS detoxification, and orchestrates the cellular antioxidant response (44). By controlling iron availability and reducing the likelihood of oxidative stress caused by iron, FTH1 suppresses the ferroptosis response (45). The enzyme GPX4 provides an important defense against iron oxidation by directly neutralizing lipid peroxides and shielding the cell membrane from oxidative stress (46). Common uses of SLC7A11 include lipid peroxidation prevention and intracellular GSH maintenance, both of which are essential for shielding cells against ferroptosis (47). In our study, we found that treatment with H2O2 not only increased the levels of iron and ROS in the MOVAS but also significantly decreased the GSH levels, highlighting the oxidative damage. NRF2, GPX4, FTH1, and SLC7A11 were among the ferroptosis inhibitors that were restored when ALAS2 overexpression reduced apoptosis and iron buildup.
In light of these findings, the role of GATA1 in aortic aneurysm pathogenesis needs to be examined further. GATA1, a crucial transcription factor known for its involvement in hematopoietic differentiation, has recently garnered attention due to its potential relevance in non-hematopoietic tissues, including the cardiovascular system. Previous studies have explored the significance of GATA1 in different disease contexts (48,49). For example, Zingariello et al. demonstrated that mice with low GATA1 expression can serve as a genuine model for myelofibrosis, as this model reflects the excessive activation of the TPO/MPL/JAK2 axis observed in the megakaryocytes of patients with bone marrow fibrosis. This study illustrated the multifaceted roles of GATA1 in regulating the critical pathways associated with hematological disorders, emphasizing the importance of this transcription factor in disease pathogenesis (50). Moreover, research by Zon et al. revealed the ability of GATA1 to activate the erythropoietin receptor promoter, underscoring its role in promoting red blood cell production (51).
The implications of these findings extended beyond erythropoiesis and encompass broader areas of hematopoiesis, reflecting the diverse functions of GATA1 in various physiological processes. To further complicate matters, our study highlights the role of ALAS2 overexpression and GATA1 knockdown in reducing H2O2-induced inflammation via pro-inflammatory cytokines, including IL-1β, TNF-α, and IL-6, exhibited a significant increase, as confirmed by previous study. These markers of inflammation highlighted the relevance of therapeutic techniques in addressing ferroptosis and inflammation, which are critical in the pathogenesis of aortic aneurysms (52).
Limitation
Our study initially explored the role of the ALAS2 gene in SMCs, and despite some findings, we need to acknowledge some limitations of the study. Our study was based on the analysis of bulk microarray datasets, which may have led to a less clear understanding of the major role factors of ALAS2 in SMCs. In order to more accurately determine the role of ALAS2 in SMCs, future studies could consider analyses using single-cell RNA sequencing data or human tissue data to provide more convincing evidence. In the discussion section, we need to explore the role of ALAS2 in more depth. Some literature reports that ALAS2 plays a negative role in muscle diseases, such as in ALAS2 overexpression mouse models. In addition, we need to consider whether ALAS2 overexpression leads to an increase in downstream HO-1 activity and explore whether this mechanism could explain the benefits of ALAS2. The future is needed to further validate the findings in this regard. We should also explicitly mention that the small haemorrhages that may have been present in the TAA tissues we observed may have partially stimulated the appearance of iron treatment in TAA. However, this is different from what may be present in cultured SMCs under experimental conditions. We need to recognize this point.
Conclusions
In summary, our study provides compelling evidence supporting the protective role of ALAS2 against oxidative stress-induced ferroptosis in MOVAS cells. Through ALAS2 overexpression, we observed a significant reduction in both iron accumulation and apoptosis triggered by H2O2 induction, underscoring its cytoprotective effects. Importantly, our investigation into the interaction between ALAS2 and GATA1 reveals a critical aspect of this defense mechanism. Specifically, we found that GATA1 knockdown reversed the cytoprotective benefits conferred by ALAS2, further emphasizing the intertwined relationship between these two factors in regulating ferroptosis and iron homeostasis. These findings not only enhance our understanding of the molecular mechanisms underlying ferroptosis but also lay the groundwork for potential therapeutic strategies aimed at mitigating ferroptosis-associated aortic aneurysm progression.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-370/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-370/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-370/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-370/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ladich E, Butany J, Virmani R. Aneurysms of the aorta: ascending, thoracic and abdominal and their management. In: Maximilian Buja L, Butany J. editors. Cardiovascular pathology. New York: Elsevier; 2016:169-211.
- Iliopoulos DC, Kritharis EP, Boussias S, et al. Biomechanical properties and histological structure of sinus of Valsalva aneurysms in relation to age and region. J Biomech 2013;46:931-40. [Crossref] [PubMed]
- Mathur A, Mohan V, Ameta D, et al. Aortic aneurysm. J Transl Int Med 2016;4:35-41. [Crossref] [PubMed]
- Sakalihasan N, Michel JB, Katsargyris A, et al. Abdominal aortic aneurysms. Nat Rev Dis Primers 2018;4:34. [Crossref] [PubMed]
- Bossone E, Eagle KA. Epidemiology and management of aortic disease: aortic aneurysms and acute aortic syndromes. Nat Rev Cardiol 2021;18:331-48. [Crossref] [PubMed]
- Kotze CW, Ahmed IG. Etiology and pathogenesis of aortic aneurysm. In: Grundmann R. editor. Etiology, pathogenesis and pathophysiology of aortic aneurysms and aneurysm rupture. Rijeka, Croatia: InTech; 2011:1-24.
- Gao J, Cao H, Hu G, et al. The mechanism and therapy of aortic aneurysms. Signal Transduct Target Ther 2023;8:55. [Crossref] [PubMed]
- Silaschi M, Byrne J, Wendler O. Aortic dissection: medical, interventional and surgical management. Heart 2017;103:78-87. [Crossref] [PubMed]
- Golledge J. Abdominal aortic aneurysm: update on pathogenesis and medical treatments. Nat Rev Cardiol 2019;16:225-42. [Crossref] [PubMed]
- Schanzer A, Oderich GS. Management of Abdominal Aortic Aneurysms. N Engl J Med 2021;385:1690-8. [Crossref] [PubMed]
- Yu Y, Yan Y, Niu F, et al. Ferroptosis: a cell death connecting oxidative stress, inflammation and cardiovascular diseases. Cell Death Discov 2021;7:193. [Crossref] [PubMed]
- Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019;12:34. [Crossref] [PubMed]
- Ren J, Lv Y, Wu L, et al. Key ferroptosis-related genes in abdominal aortic aneurysm formation and rupture as determined by combining bioinformatics techniques. Front Cardiovasc Med 2022;9:875434. [Crossref] [PubMed]
- Filiberto AC, Ladd Z, Leroy V, et al. Resolution of inflammation via RvD1/FPR2 signaling mitigates Nox2 activation and ferroptosis of macrophages in experimental abdominal aortic aneurysms. FASEB J 2022;36:e22579. [Crossref] [PubMed]
- Loick P, Mohammad GH, Cassimjee I, et al. Protective Role for Smooth Muscle Cell Hepcidin in Abdominal Aortic Aneurysm. Arterioscler Thromb Vasc Biol 2023;43:713-25. [Crossref] [PubMed]
- Poli A, Schmitt C, Moulouel B, et al. Iron, Heme Synthesis and Erythropoietic Porphyrias: A Complex Interplay. Metabolites 2021;11:798. [Crossref] [PubMed]
- Dutt S, Hamza I, Bartnikas TB. Molecular Mechanisms of Iron and Heme Metabolism. Annu Rev Nutr 2022;42:311-35. [Crossref] [PubMed]
- Sawicki KT, Shang M, Wu R, et al. Increased Heme Levels in the Heart Lead to Exacerbated Ischemic Injury. J Am Heart Assoc 2015;4:e002272. [Crossref] [PubMed]
- Pilling LC, Joehanes R, Kacprowski T, et al. Gene transcripts associated with muscle strength: a CHARGE meta-analysis of 7,781 persons. Physiol Genomics 2016;48:1-11. [Crossref] [PubMed]
- Peng Y, Li J, Luo D, et al. Muscle atrophy induced by overexpression of ALAS2 is related to muscle mitochondrial dysfunction. Skelet Muscle 2021;11:9. [Crossref] [PubMed]
- Reinwald H, Alvincz J, Salinas G, et al. Toxicogenomic profiling after sublethal exposure to nerve- and muscle-targeting insecticides reveals cardiac and neuronal developmental effects in zebrafish embryos. Chemosphere 2022;291:132746. [Crossref] [PubMed]
- Massaiu I, Campodonico J, Mapelli M, et al. Dysregulation of Iron Metabolism-Linked Genes at Myocardial Tissue and Cell Levels in Dilated Cardiomyopathy. Int J Mol Sci 2023;24:2887. [Crossref] [PubMed]
- Virmani R, Sato Y, Sakamoto A, et al. Aneurysms of the aorta: ascending, thoracic, and abdominal and their management. In: Maximilian Buja L, Butany J. editors. Cardiovascular Pathology. New York: Elsevier; 2022:353-406.
- Harky A, Singh VP, Khan D, et al. Factors Affecting Outcomes in Acute Type A Aortic Dissection: A Systematic Review. Heart Lung Circ 2020;29:1668-81. [Crossref] [PubMed]
- Habets J, Zandvoort HJ, Reitsma JB, et al. Magnetic resonance imaging is more sensitive than computed tomography angiography for the detection of endoleaks after endovascular abdominal aortic aneurysm repair: a systematic review. Eur J Vasc Endovasc Surg 2013;45:340-50. [Crossref] [PubMed]
- Frazao C, Tavoosi A, Wintersperger BJ, et al. Multimodality Assessment of Thoracic Aortic Dimensions: Comparison of Computed Tomography Angiography, Magnetic Resonance Imaging, and Echocardiography Measurements. J Thorac Imaging 2020;35:399-406. [Crossref] [PubMed]
- Stilo F, Catanese V, Nenna A, et al. Biomarkers in EndoVascular Aneurysm Repair (EVAR) and Abdominal Aortic Aneurysm: Pathophysiology and Clinical Implications. Diagnostics (Basel) 2022;12:183. [Crossref] [PubMed]
- Xue C, Yang B, Fu L, Hou H, Qiang J, Zhou C, et al. Urinary biomarkers can outperform serum biomarkers in identifying certain diseases. URINE 2023;
- Alamoudi AO. The influence of different diagnostic imaging and interventional repair techniques on mortality rate in aortic aneurysm patients. Rutgers University-School of Health Related Professions; 2015. Doi:
10.7282/T31G0P66 . - Comparative clinical effectiveness and cost effectiveness of endovascular strategy v open repair for ruptured abdominal aortic aneurysm: three year results of the IMPROVE randomised trial. BMJ 2017;359:j4859. [Crossref] [PubMed]
- Alsawas M, Zaiem F, Larrea-Mantilla L, et al. Effectiveness of surgical interventions for thoracic aortic aneurysms: A systematic review and meta-analysis. J Vasc Surg 2017;66:1258-1268.e8. [Crossref] [PubMed]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266-82. [Crossref] [PubMed]
- Ono K, Fujiwara T, Saito K, et al. Congenital sideroblastic anemia model due to ALAS2 mutation is susceptible to ferroptosis. Sci Rep 2022;12:9024. [Crossref] [PubMed]
- Lv P, Liu F. Heme-deficient primitive red blood cells induce HSPC ferroptosis by altering iron homeostasis during zebrafish embryogenesis. Development 2023;150:dev201690. [Crossref] [PubMed]
- Whatley SD, Ducamp S, Gouya L, et al. C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am J Hum Genet 2008;83:408-14. [Crossref] [PubMed]
- Song J, Nilsson G, Xu Y, et al. Temporal brain transcriptome analysis reveals key pathological events after germinal matrix hemorrhage in neonatal rats. J Cereb Blood Flow Metab 2022;42:1632-49. [Crossref] [PubMed]
- Totsuka K, Ueta T, Uchida T, et al. Oxidative stress induces ferroptotic cell death in retinal pigment epithelial cells. Exp Eye Res 2019;181:316-24. [Crossref] [PubMed]
- Park MW, Cha HW, Kim J, et al. NOX4 promotes ferroptosis of astrocytes by oxidative stress-induced lipid peroxidation via the impairment of mitochondrial metabolism in Alzheimer's diseases. Redox Biol 2021;41:101947. [Crossref] [PubMed]
- Fujii J, Homma T, Kobayashi S. Ferroptosis caused by cysteine insufficiency and oxidative insult. Free Radic Res 2020;54:969-80. [Crossref] [PubMed]
- Zhou M, Zha Z, Zheng Z, et al. Cordycepin suppresses vascular inflammation, apoptosis and oxidative stress of arterial smooth muscle cell in thoracic aortic aneurysm with VEGF inhibition. Int Immunopharmacol 2023;116:109759. [Crossref] [PubMed]
- Branchetti E, Poggio P, Sainger R, et al. Oxidative stress modulates vascular smooth muscle cell phenotype via CTGF in thoracic aortic aneurysm. Cardiovasc Res 2013;100:316-24. [Crossref] [PubMed]
- Portelli SS, Hambly BD, Jeremy RW, et al. Oxidative stress in genetically triggered thoracic aortic aneurysm: role in pathogenesis and therapeutic opportunities. Redox Rep 2021;26:45-52. [Crossref] [PubMed]
- Sharma A, Flora SJS. Positive and Negative Regulation of Ferroptosis and Its Role in Maintaining Metabolic and Redox Homeostasis. Oxid Med Cell Longev 2021;2021:9074206. [Crossref] [PubMed]
- He F, Ru X, Wen T. NRF2, a Transcription Factor for Stress Response and Beyond. Int J Mol Sci 2020;21:4777. [Crossref] [PubMed]
- Song L, Wang X, Cheng W, et al. Expression signature, prognosis value and immune characteristics of cathepsin F in non-small cell lung cancer identified by bioinformatics assessment. BMC Pulm Med 2021;21:420. [Crossref] [PubMed]
- Lin KJ, Chen SD, Lin KL, et al. Iron Brain Menace: The Involvement of Ferroptosis in Parkinson Disease. Cells 2022;11:3829. [Crossref] [PubMed]
- Wang L, Liu Y, Du T, et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc Cell Death Differ 2020;27:662-75. [Crossref] [PubMed]
- Tsiftsoglou AS. Erythropoietin (EPO) as a Key Regulator of Erythropoiesis, Bone Remodeling and Endothelial Transdifferentiation of Multipotent Mesenchymal Stem Cells (MSCs): Implications in Regenerative Medicine. Cells 2021;10:2140. [Crossref] [PubMed]
- Anzai A, Ko S, Fukuda K. Immune and Inflammatory Networks in Myocardial Infarction: Current Research and Its Potential Implications for the Clinic. Int J Mol Sci 2022;23:5214. [Crossref] [PubMed]
- Zingariello M, Sancillo L, Martelli F, et al. The thrombopoietin/MPL axis is activated in the Gata1(low) mouse model of myelofibrosis and is associated with a defective RPS14 signature. Blood Cancer J 2017;7:e572. [Crossref] [PubMed]
- Zon LI, Youssoufian H, Mather C, et al. Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc Natl Acad Sci U S A 1991;88:10638-41. [Crossref] [PubMed]
- Batra R, Suh MK, Carson JS, et al. IL-1β (Interleukin-1β) and TNF-α (Tumor Necrosis Factor-α) Impact Abdominal Aortic Aneurysm Formation by Differential Effects on Macrophage Polarization. Arterioscler Thromb Vasc Biol 2018;38:457-63. [Crossref] [PubMed]


