
This article has an erratum available at: http://dx.doi.org/10.21037/jtd-2025b-01 the article has been update on 2025-01-16 at here.
KLRB1 expression is associated with lung adenocarcinoma prognosis and immune infiltration and regulates lung adenocarcinoma cell proliferation and metastasis through the MAPK/ERK pathway
Highlight box
Key findings
• Studies have shown that KLRB1 is associated with lung adenocarcinoma prognosis and immune infiltration and is a novel biomarker in lung adenocarcinoma.
What is known, and what is new?
• We know little about KLRB1 in terms of maintaining malignant phenotype and immune infiltration in lung cancer.
• High KLRB1 expression is associated with a favorable prognosis in most cancers.
What is the implication, and what should change now?
• KLRB1 expression is linked to prognosis, immune infiltration, and migration.
• KLRB1 expression is linked to proliferation in lung adenocarcinoma.
Introduction
Lung cancer is a frequently occurring form of cancer and is responsible for the largest number of cancer-related fatalities globally (1). The incidence of lung adenocarcinoma (LUAD), the predominant histological subtype of lung cancer, continues to increase among individuals who currently smoke, previously smoked, or never smoked (2).
Despite significant progress in our understanding of the disease pathogenesis and the development of innovative therapies, the cure rate of lung cancer therapies remains unsatisfactory.
The KLRB1 gene is responsible for encoding the CD161 receptor found on natural killer (NK) cells. It is a C-type lectin-like receptor found on the surface of NK cells and certain subsets of T cells (3-6) that plays a crucial role in controlling NK cell cytotoxicity and promoting T cell proliferation through interaction with its ligand LLT1. IFN-γ and IL-17 are secreted through the interaction between the ligand and receptor (7-9). Moreover, the correlation of CD161 with T-cell stimulation is supported by certain lines of evidence. Costimulation and differentiation are essential processes in cellular biology. CD161 may be actively involved in tumor immunosurveillance (10). Single-cell RNA sequencing of tumor-infiltrating T cells identified CD161 as a potential inhibitory receptor in gliomas (11). CD8+PD-1+CD161+ T-cell subsets are more cytotoxic and proliferative in hepatocellular carcinoma (HCC) than in other cancer types (12). These results suggested that CD161 plays an important role in tumor immune regulation. Previous studies have shown that KLRB1 transcription is repressed in tumor tissue in 68% of non-small cell lung cancer patients and that CD161 expression in lung cancer is associated with improved clinical outcomes (3,10). However, the potential effect of KLRB1 on LUAD and its relationship with tumor-infiltrating immune cells (TIICs) are not fully understood.
The aim of this study was to investigate the relationship between KLRB1 expression and clinicopathological prognosis, potential molecular mechanisms, and tumor immunity using bioinformatics analysis to help clinicians refine treatment and improve the prognosis of lung cancer patients. We present this article in accordance with the MDAR and TRIPOD reporting checklists (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-8/rc).
Methods
Clinical specimens and data collection and processing
The Department of Oncology, Huashan Hospital, Fudan University, supplied us with 72 sets of lung cancer patient samples for clinical research. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients in our study provided consent after being fully informed. Our research was approved by the ethics committee of Huashan Hospital, Fudan University (2022 Pro Review No. 634). The RNA-seq data in fragments per kilobase per million (FPKM) format and clinicopathological information of 594 LUAD patients were obtained for The Cancer Genome Atlas lung adenocarcinoma cohort (TCGA-LUAD) via the TCGA website (https://portal.gdc.cancer.gov/). The FPKM format of the RNA sequencing data was converted to TPM format, which represents the transcripts per million reads. For the purpose of our analysis, we obtained four LUAD datasets (GSE116959, GSE27262, GSE10072, and GSE33532) from the GEO database website (https://www.geo.ncbi.nlm.nih.gov/.ncbi.nlm.nih.gov/geo/). The TCGA and GEO databases offer publicly accessible data. The expression levels of KLRB1 mRNA were determined through analysis of 535 LUAD tissue samples and 59 normal lung tissue samples obtained from the TCGA database. KLRB1 expression in the LUAD samples, which included samples from the GSE116959, GSE27262, GSE10072, and GSE33532 datasets, was analyzed using the GEO database.
Differentially expressed gene (DEG) analysis
By considering the median value of KLRB1 expression, the expression levels of KLRB1 were used to stratify patients with lung cancer in the TCGA dataset into two groups: the high expression group and the low expression group. Subsequently, the analysis of DEGs between these groups was carried out using the DESeq2 R package (13), and volcano plots and heatmaps of the DEGs were generated using the R package “ggplot2” (v3.3.3), with a log-fold change threshold of absolute value 1 and a P value threshold of 0.05. Correlations between the expression of DEGs and KLRB1 were assessed using Spearman correlation analysis and visualized with ggplot2 [version 3.3.3].
Functional enrichment analysis
The “org.Hs.eg.db” (v3.10.0) R package was used to convert the Entrez ID to a gene symbol. Functional enrichment analyses, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, were implemented for the DEGs using the R package “ClusterProfiler” (v3.14.3) and visualized via the R package “ggplot2” (14). Gene set enrichment analysis (GSEA) was carried out using the R package “ClusterProfiler” (v3.14.3), and functional or pathway terms with an adjusted P value <0.05 and a false discovery rate (FDR) <0.25 were regarded as significantly enriched (14,15).
Evaluation of the prognostic significance of KLRB1 expression in LUAD
The researchers utilized the Kaplan-Meier method with the log-rank test to conduct survival analysis. Using the “surv-cutpoint” function from the survminer R package, patients were categorized into high and low KLRB1 expression groups by evaluating all possible cutoff points to determine the optimal segmentation. Statistical analysis was performed to calculate and analyze the hazard ratio (HR), 95% confidence interval (95% CI), and log-rank P value. Univariate and multivariate Cox regression analyses were used to evaluate how clinical factors affect patient outcomes. Column line plots were used to derive independent prognostic factors in multivariate Cox analysis for predicting the overall survival (OS) probability. Using the R package RMS (v6), line plots were generated and organized into columns. Column line plots were constructed using the R package RMS (v6.2-0). Time-dependent survival (receptor operating characteristic, ROC) curves were analyzed using “pROC” (v1.17.0.1), “timeROC” (v0.4) and “ggplot2” (v3.3.3) to evaluate the predictive value of KLRB1 expression levels in the diagnosis of LUAD. ROC curves were used to assess the predictive value of KLRB1 expression levels for LUAD diagnosis. Kaplan-Meier survival curves were used to analyse the prognostic value of KLRB1 in a subgroup of LUAD patients.
Analysis of immune infiltration and its relationship with immunomodulators
The tumor infiltration status of 24 immune cell types was assessed using the single-sample GSEA (ssGSEA) algorithm of the “GSVA” (v1.34.0) R package (16,17). We conducted a Spearman correlation analysis to assess the relationship between the expression of KLRB1 and these immune cells. To gain further insight into the association between KLRB1 expression and immunomodulators, we utilized the TISIDB database (http://cis.hku.hk/TISIDB) (18). Through Spearman correlation analysis, we assessed the significant associations between KLRB1 expression and both immunosuppressors and immunostimulators. Next, we developed prognostic multi-immune genetic signatures based on KLRB1-associated immunomodulators. Then, they were processed with the least absolute shrinkage and selection operator (LASSO) to avoid overfitting and eliminate closely related genes. Tenfold cross-validation was used to select the minimum penalty term (λ). After that, prognostic models of immune molecules associated with KLRB1 were constructed. Risk score = β1x1 + β2x2 + ... + βixi; in this formula, xi represents the expression of each gene, and βi represents the coefficient of each gene.
Cell culture
A549 human lung cancer cells were from the American Type Culture Collection (ATCC). The cells were cultured in RPMI-1640 culture medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Yeasen Biotech, Shanghai, China) and 1% penicillin-streptomycin (Sigma-Aldrich, St. Louis, MO, USA) and then maintained in an incubator at a constant temperature of 37 ℃ and a humidified atmosphere with 5% CO2.
Cell transfection
The cells were seeded onto a 6-well plate. Small interfering RNAs (siRNAs) against human KLRB1 were transfected into the human LUAD cell Line A549 with Lipofectamine 2000 according to the instructions (Invitrogen, Carlsbad, CA, USA). Forty-eight hours after cell transfection, the cells were collected, and the interference efficiency was assessed by Western blotting. Three siRNAs directed against KLRB1 mRNA were synthesized by RiboBio (Guangzhou, China).
Migration and invasion assays
Migration and invasion assays were performed in 24-well plates with 8 mm pore size chamber inserts (Corning, NY, USA). In the invasion assay, the upper chamber was coated with Matrigel to simulate the vascular basement membrane. However, in the migration assay, the chambers were placed in 24-well plates without Matrigel. A total of 1×105 and 5×104 lung cancer cells were resuspended in 200 µL of serum-free medium and added to the upper chamber, which was then placed into a 24-well plate for invasion and migration assays, respectively. Then, 800 µL of medium supplemented with 10% FBS was added to the lower chamber as a chemical stimulant for 18 hours. Cells that traversed the membrane (traversal) or matrix gel (invasion) were chemically fixed for 30 minutes using 100% methanol and subsequently stained with crystal violet dye for 15 minutes. The number of cells present in nine randomly selected microscopic areas was then determined under a microscope.
Cell proliferation assay
In the proliferation assay, we inoculated 800 control and treated lung cancer cells into 96-well plates. Twenty-four hours later, we measured the optical density (OD) 450 of each well for 5 consecutive days using a Cell Counting Kit-8 (CCK-8) assay (Dojindo Corp, Tokyo, Japan). According to the protocol, the medium in the original culture wells was first discarded, and then a mixture of 10 µL of reagent and 90 µL of medium provided in the kit was added to each well. After 2 hours of incubation, we quantified the degree of light absorption at a wavelength of 450 nm. Eventually, a growth curve was constructed using the measured OD at 450 nm. In the colony formation assay, 500 cells were inoculated into 6-well plates. The cells were incubated in an incubator with 2.5 mL of culture medium at 37 ℃. After 7 days of incubation, the cells were carefully rinsed with phosphate-buffered saline (PBS). Each well was treated with 2 mL of pure methanol, ensuring that the colonies were effectively fixed for 15 minutes. After 20 minutes, crystal violet solution was added for staining. Microscopic imaging was employed for the purpose of quantification.
Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR)
RNA was extracted from all cell samples used in our study using TRIzol reagent (Invitrogen). cDNA was synthesized with PrimeScript RT Master Mix (TaKaRa, Tokyo, Japan). cDNA was used as a template for qPCR with SYBR Green Premix Ex Tag (TaKaRa). The following primers were used in this study: KLRB1 forward (5'-TGGCATCAATTTGCCCTGAAA-3') and reverse (5'-TCCAAGGGTTGACAGTGTGAG-3'). β-actin: forward (5'-GTCATTCCAAATATGAGATGCG-3') and reverse (5'-GCATTACATAATTTACACGAAAGCA-3'). β-actin was used as an internal control. Relative expression differences were calculated using the 2−ΔΔCt method.
Protein blotting assay
Proteins were extracted from the samples and cells using T-PER® Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with phosphatase inhibitor and protease inhibitor cocktails (Yeasen Biotech). After the proteins were quantified by a BCA protein assay, the proteins obtained were loaded on SDS polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The PVDF membranes were blocked with 5% skim milk in PBST for one hour and then incubated with the following primary antibodies at 4 ℃ overnight: anti-KLRB1 (1:500, 67537-1-Ig, Proteintech, Wuhan, China); anti-Raf-1 (1:500, BD-PT3979, Biodragon, Wuhan, China); anti-pRaf-1 (1:500, BD-PP0516, Biodragon); anti-MEK-1/2 (1:500, BD-PT2715, Biodragon); anti-pMEK-1/2 (1:500, BD-PP0167, Biodragon); anti-ERK1/2 (1:500, BD-PT1625, Biodragon); anti-pERK1/2 (1:1,000, #4370, CST, Danvers, MA, USA); anti-MSK1 (1:500, BD-PT2902, Biodragon); anti-pMSK1 (1:500, BD-PP0517, Biodragon); anti-vinculin (1:3,000, #4650, CST); and anti-β-actin (1:5,000, HRP-66009, Proteintech). After washing 3 times with 0.1% PBST, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (CST) for 1.5 hours. The bands were visualized using a chemiluminescence system (Bio-Rad, Hercules, CA, USA), and quantitative analysis was performed using Quantity One software (Bio-Rad).
Statistical analysis
Statistical analyses to determine the significance of KLRB1 expression in unpaired and paired tissues were conducted using the bioinformatics online database and R (version 3.6.3), employing the Weltch t-test and paired samples t-test. The Wilcoxon rank sum test was used to determine if there were significant differences between two groups. With respect to accurate detection in fish, the chi-squared test and logistic regression were used to evaluate the associations between clinical attributes and KLRB1 expression. Both statistical tests were conducted using a two-tailed approach, and P values <0.05 were regarded as statistically significant.
Results
KLRB1 expression levels in LUAD and clinical correlation analysis
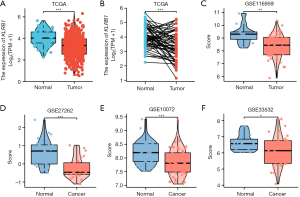
TCGA database analysis revealed that KLRB1 was expressed at low levels in LUAD patients (P<0.001) (Figure 1A). We also verified that the expression of KLRB1 in LUAD tissues was notably lower than that in the corresponding normal tissues (P<0.001) (Figure 1B). Utilizing the GEO datasets (GSE116959, GSE27262, GSE10072, and GSE33532), the expression of KLRB1 was assessed in lung cancer. There was a noteworthy decrease in KLRB1 levels in lung cancer tissues compared to normal lung tissues (Figure 1C-1F).
The associations between clinicopathological characteristics and KLRB1 expression levels in patients with LUAD based on the TCGA-LUAD dataset are shown in Figure S1. Significant differences in clinical T stage, M stage, age, sex, smoking status, pathologic stage, first-line treatment outcome, OS, and disease-specific survival (DSS) were observed between LUAD patients with high and low KLRB1 expression. Moreover, univariate logistic regression analyses revealed differences in some clinicopathological features including T stage (T2, T3, and T4 vs. T1), age (>65 vs. ≤65 years), smoking status (yes vs. no), and first-line treatment outcome [progressive disease (PD), stable disease (SD), partial response (PR) vs. complete response (CR)] between the groups with high and low KLRB1 expression (Table 1). The findings revealed that LUAD patients who exhibited low KLRB1 expression levels had a greater likelihood of presenting with malignant phenotypes.
Table 1
| Characteristics | Total (N) | OR (95% CI) | P value |
|---|---|---|---|
| T stage (T2 & T3 & T4 vs. T1) | 510 | 0.452 (0.308–0.658) | <0.001* |
| N stage (N1 & N2 & N3 vs. N0) | 501 | 0.709 (0.488–1.027) | 0.07 |
| M stage (M1 vs. M0) | 369 | 0.465 (0.185–1.075) | 0.08 |
| Pathologic stage (III & IV vs. I & II) | 505 | 0.653 (0.424–0.998) | 0.05 |
| Gender (male vs. female) | 513 | 0.736 (0.519–1.043) | 0.09 |
| Age (>65 vs. ≤65 years) | 494 | 1.896 (1.328–2.717) | <0.001* |
| Race (Black or African American vs. Asian & White) | 446 | 0.606 (0.331–1.088) | 0.10 |
| Smoker (yes vs. no) | 499 | 0.551 (0.329–0.910) | 0.02* |
| Pack-year (≥40 vs. <40) | 351 | 0.844 (0.554–1.282) | 0.43 |
| Residual tumor (R1 & R2 vs. R0) | 361 | 1.921 (0.714–5.683) | 0.21 |
| Anatomic neoplasm subdivision (right vs. left) | 498 | 0.920 (0.642–1.317) | 0.65 |
| Primary therapy outcome (PD & SD & PR vs. CR) | 426 | 0.620 (0.398–0.959) | 0.03* |
*, two-sided P<0.05. PD, progressive disease; SD, stable disease; PR, partial response; CR, complete response.
DEGs between LUAD patients with high and low KLRB1 expression
The 594 patients diagnosed with LUAD were categorized into two groups based on the median expression value of the KLRB1 gene. A total of 927 genes were differentially expressed between the KLRB1 groups with high and low expression, with 529 genes upregulated and 398 genes downregulated [adjusted P value <0.05, |log2fold change (FC)| >1] (Figure 2A; the supplementary table is available at https://cdn.amegroups.cn/static/public/jtd-24-8-1.xlsx). Figure 2B,2C illustrates the top 20 DEGs that were positively and negatively correlated, as indicated by the single-gene correlation heatmap.

Functional enrichment analysis of KLRB1-associated DEGs in LUAD
We used the “clusterProfiler” R package for functional annotation of KLRB1-related DEGs in LUAD patients. The results of GO enrichment analysis, including highly enriched biological processes, cellular components and molecular functions (P<0.05), are illustrated in Figure 2D and in the supplementary table (https://cdn.amegroups.cn/static/public/jtd-24-8-2.xlsx).
The primary biological processes included the activation of T cells, the regulation of lymphocyte activation, the regulation of T-cell activation, the differentiation of lymphocytes, and the differentiation of T cells. The cellular components that exhibited the highest levels of enrichment were the external side of plasma membrane, complexes of MHC class II proteins, immunological synapses, complexes of MHC proteins, and neuronal cell bodies. In the dataset, the predominant molecular functions were cytokine receptor activity, C-C chemokine binding, cytokine activity, G protein-coupled chemoattractant receptor activity and chemokine receptor activity. Moreover, KEGG pathway analysis revealed significant enrichment of DEGs related to various pathways, including interactions between cytokines and cytokine receptors, interactions between cell adhesion molecules, differentiation of Th17 cells, differentiation of Th1 and Th2 cells, signaling pathways of chemokines, signaling pathways of T-cell receptors, cytotoxicity mediated by NK cells, and cascades involving complement and coagulation (Figure 2E; the supplementary table is available at https://cdn.amegroups.cn/static/public/jtd-24-8-3.xlsx). Then, the KLRB1 high- and low-expression groups were compared using GSEA. GSEA revealed that the KLRB1 high-expression group exhibited significant enrichment of genes related to immune-related biological processes, such as NK cell-mediated immunity, T-cell chemotaxis, leukocyte-mediated cytotoxicity, lymphocyte-mediated immunity, B-cell-mediated immunity, the T-cell receptor complex, the immunoglobulin complex, and immune synapses (Figure 3A-3C; the supplementary table is available at https://cdn.amegroups.cn/static/public/jtd-24-8-4.xlsx). Similarly, the KEGG pathway analysis demonstrated notable enrichment of genes involved in various biological processes, including cytokine and cytokine receptor interactions, antigen processing and presentation, NK cell-mediated cytotoxicity, the T-cell receptor signaling pathway, the B-cell receptor signaling pathway, leukocyte transendothelial migration, and the JAK-STAT signaling pathway (Figure 3D; the supplementary table is available at https://cdn.amegroups.cn/static/public/jtd-24-8-4.xlsx). These findings indicated that low KLRB1 expression might be associated with an immunosuppressive phenotype.

One particular change that attracted our attention was the increase in MAPK/ERK cascade activity (Figure 4A). Hence, we assessed the potential involvement of KLRB1 in the regulation of this pathway. The phosphorylation of RAF-1, MEK-1/2, ERK1/2 and MSK1 was augmented after KLRB1 knockdown, as shown by protein blotting analysis (Figure 4B). These findings suggest that inhibition of KLRB1 can enhance the proliferation and growth of LUAD cells via the MAPK/ERK signaling pathway.

Correlation between KLRB1 expression and immune infiltration
The infiltration status of 24 immune cell types in LUAD tissues was assessed by ssGSEA, and Spearman correlation analysis was performed to assess the correlation between KLRB1 expression and immune cell infiltration. KLRB1 expression was positively correlated with the levels of T cells (r=0.873, P<0.001), cytotoxic cells (r=0.777, P<0.001), B cells (r=0.661, P<0.001), and Th1 cells (r=0.639, P<0.001) (Figure S2A). The tumor infiltration levels of T cells, cytotoxic cells, B cells and Th1 cells were consistent with the results of Spearman’s analysis (Figure S2B-S2I).
Effect of KLRB1-related immunomodulators on the prognosis of LUAD
We identified 39 immunostimulators (C10orf54, CD27, CD276, CD28, CD40, CD40LG, CD48, CD70, CD80, CD86, CXCL12, CXCR4, ENTPD1, HHLA2, ICOS, ICOSLG, IL2RA, IL6, IL6R, KLRC1, KLRK1, LTA, MICB, PVR, TMEM173, TMIGD2, TNFRSF13B, TNFRSF13C, TNFRSF14, TNFRSF17, TNFRSF18, TNFRSF4, TNFRSF8, TNFRSF9, TNFSF13, TNFSF13B, TNFSF14, TNFSF15 and TNFSF4) (Figure 5A) and 20 immunosuppressive factors (ADORA2A, BTLA, CD160, CD244, CD274, CD96, CSF1R, CTLA4, HAVCR2, IDO1, IL10, IL10RB, LAG3, LGALS9, PDCD1, PDCD1LG2, PVRL2, TGFB1, TGFBR1 and TIGIT) (Figure 5B). The levels of these genes were significantly associated with those of KLRB1 in LUAD. One-way Cox regression analysis was conducted to investigate the predictive significance of KLRB1-related immunomodulators in LUAD. We identified 16 genes that were significantly associated with prognosis in LUAD patients (Figure 5C). After that, a four-gene prognostic model was developed using LASSO regression analysis (Figure 5D,5E). Risk assessments were conducted by summing the multiplicative product of expression values and coefficients for each gene, enabling the prediction of potential risks. In the context of the study, the analysis of Kaplan-Meier survival curves revealed a pronounced disparity in survival durations between patients with high risk scores and those with low risk scores; Survival time was significantly shorter in the former group (P<0.001) (Figure 5F). In addition, we investigated the relationship between risk score and survival status in LUAD patients, and the expression patterns of four genes in the high- and low-risk groups are shown in the heatmap (Figure 5G).

Prognostic value of KLRB1 in lung cancer
Kaplan-Meier survival curve analysis revealed that LUAD patients with low KLRB1 expression had markedly reduced OS (P=0.005) (Figure 6A) and DSS (P=0.008) (Figure 6B). Next, the associations between the expression of KLRB1 and the predicted outcomes in diverse subcategories were assessed. Regardless of OS or DSS, patients with low KLRB1 expression, including those in various subgroups (T1, T2, N0, M0, <40 pack years, smokers, females, age >65 years, stage I disease and stage II disease), had notably worse prognoses than patients with high KLRB1 expression (all P<0.05) (Figure S3).

A comprehensive analysis including both univariate and multivariate Cox regression analyses was conducted to identify factors influencing prognosis. Univariate Cox analysis revealed that T stage (adjusted HR =2.317, 95% CI: 1.591–3.375, P<0.001), N stage (adjusted HR =2.321, 95% CI: 1.631–3.303, P<0.001), M stage (adjusted HR =2.136, 95% CI: 1.248–3.653, P=0.006), and KLRB1 (adjusted HR =0.797, 95% CI: 0.691–0.920, P=0.002) were associated with OS in LUAD patients (Figure 6C). Multivariate analysis revealed that KLRB1 expression (adjusted HR =0.833, 95% CI: 0.703–0.986, P=0.03), T stage (adjusted HR =1.826, 95% CI: 1.192–2.795, P=0.006), and N stage (adjusted HR =2.041, 95% CI: 1.359–3.065, P<0.001) were independent prognostic factors for OS in lung cancer patients (Figure 6D). Similarly, for DSS, KLRB1 expression (adjusted HR =0.754, 95% CI: 0.600–0.946, P=0.02), M stage (adjusted HR =2.162, 95% CI: 1.069–4.370, P=0.03), and T stage (adjusted HR =1.758, 95% CI: 1.027–3.007, P=0.04) were shown to be prognostic factors (Table S1).
A comprehensive evaluation of KLRB1 expression levels was carried out using ROC curve analysis to determine its diagnostic significance. The ROC curve showed that KLRB1 expression had good predictive ability, and the area under the curve (AUC) was 0.724, indicating that KLRB1 could distinguish between lung cancer tissues and normal tissues (Figure S4A). Time-dependent ROC curve analysis revealed that the AUC values of the 1-, 3-, and 5-year survival rates of LUAD patients predicted based on KLRB1 expression levels were 0.418, 0.427, and 0.449, respectively (Figure S4B). A column line graph model was constructed, which included T stage, N stage, and KLRB1 expression level as parameters. Multivariate Cox regression analysis revealed that these factors were highly significant prognostic factors. Columnar plots demonstrated the significant clinical value in terms of predicting survival rates for patients with LUAD over the course of 1, 3, and 5 years (Figure S4C).
Validation of KLRB1 expression levels in clinical lung cancer samples
We assessed the expression of KLRB1 in 72 pairs of cancer tissue samples and matched noncancer tissue samples that were collected in our laboratory using RT-qPCR (Figure 7A,7B) and in 15 pairs of lung cancer tissue samples and their matched noncancer tissue samples using Western blotting (Figure 7C-7F). Analysis revealed significantly lower expression of KLRB1 at the mRNA and protein levels in cancer tissues than in paracancerous tissues.

Silencing KLRB1 promotes the migration, invasion and proliferation of LUAD cells in vitro
After generating three RNA interference fragments, we examined the involvement of KLRB1 in lung cancer. RT-qPCR and Western blotting indicated that the siRNA suppressed the expression of KLRB1 in A549 cells (Figure 8A,8B). Based on these findings, we assessed the migratory and invasive capacities of A549 cells in the control group (NC group) and the group with KLRB1 knockdown (knockdown group). The results indicated a notable increase in the migration and invasion capacities of the KLRB1-si1 and KLRB1-si3 groups compared to those of the control group (Figure 8C). These findings suggested that in vitro, the absence of KLRB1 increased the metastatic potential of lung cancer cells. The CF assay was used to determine the capacity of KLRB1 to form colonies. The data revealed a profound enhancement in the CF capacity of A549 cells with reduced levels of KLRB1 (Figure 8D). The viability of A549 cells was significantly increased when KLRB1 was knocked down, as demonstrated by the CCK-8 assay (Figure 8E).

Discussion
There are many examples of similar bioinformatics studies revealing the impact of biomarkers on tumors. The statistical correlation between SYTL1 expression and endometrial cancer (EC) clinical prognosis, DNA methylation and immune cell infiltration demonstrated that SYTL1 can be used as a promising diagnostic and prognostic biomarker in EC (19). High COMMD10 expression is significantly associated with a poor prognosis in patients with gastric cancer (STAD), and its functional realization is related to m6A modification. COMMD10 is involved in the regulation of immune infiltration in STAD and promotes the generation of an immunosuppressive phenotype (20). Another study investigated the relationship between the lncRNA TRHDE-AS1 and the development of gliomas. The prognostic analysis, differential expression analysis, potential pathway mechanism analysis, mutation analysis and immune infiltration analysis of the expressed genes indicated that this lncRNA has great potential in guiding clinical prognosis prediction and future therapeutic decisions (21). According to database analysis, INHA is a diagnostic and prognostic marker for LUAD, and high expression of this molecule is a risk factor for a poor prognosis in LUAD patients and promotes LUAD cell proliferation and invasion in vitro and in vivo by activating the EGFR pathway (22). ULBP2 is an immune marker and a prognostically relevant biomarker for colon cancer (CC) patients. In CC, ULBP2 forms a complex immunosuppressive tumor microenvironment (TME) by altering infiltrating immune cell subpopulations and thus may be a potential target for CC immunotherapy (23). Compared to single omics, multiomics involves the integration of information from multiple genomics, proteomics, metabolomics, transcriptomics, etc., to explore the roles of various genomics methods in jointly influencing the development of diseases. With the development of sequencing technology and bioinformatics analysis, the framework of integrating spatiotemporal multiomics and systems biology is expected to become a universal template for analyzing complex pathobiology in various disease states. Multiomics analysis can help predict the effect of tumor immunotherapy and reveal the mechanism by which different histologies jointly influence the body’s immune system. A study reveals that metabolism influences the immune microenvironment of melanoma and predicts the response of tumor patients to anti-PD-1 immune checkpoint blockade therapy by combining different metabolic components, such as glucose metabolism, lipid metabolism, and amino acid metabolism (24). The novel multiomics integration strategy used, including transcriptomics, proteomics, and metabolomics, successfully identified four gastric cancer populations with different metabolic profiles, which will aid in the development of more targeted gastric cancer (GC) treatments (25). Multiomics data can also be used to screen for specific tumor diagnostic biomarkers. For example, analysis of the molecular profiles of samples based on different types of histological analyses is important for identifying novel biomarkers for ovarian cancer (OC) and improving clinical outcomes (26). Compared with traditional biomarkers, high-throughput multiomics technology provides new insights and a mechanistic understanding of HCC, enabling the development of all-in-one biomarkers for diagnosis (27). Thus, the development of multiomics may provide new insights into tumor-intrinsic precision medicine.
KLRB1 is located on human chromosome 12 and encodes CD161 that is expressed on the surface of most NK cells and T lymphocytes (4). Studies have shown that inhibition of KLRB1 expression in breast invasive carcinoma is associated with impaired tumor immune function, leading to tumor progression and poor prognosis (28,29). Downregulation of KLRB1 expression is strongly associated with poor survival outcomes in EC and can influence tumor progression by affecting immune cell infiltration subpopulations in the TME (30). KLRB1 exhibits potent anti-tumor immunity in human papillomavirus (HPV)-positive oropharyngeal squamous cell carcinoma (OPSCC), and high levels of intratumoral CD161 cytotoxic T lymphocytes (CTLs) are associated with favorable treatment response and prolonged OS (31). In HCC, KLRB1 is negatively correlated with immune infiltration and is associated with better OS in HCC (32). In LUAD, accumulation of KLRB1+CD8+ T cells significantly correlates with a favorable prognosis, but KLRB1+CD8+ T cell infiltration is reduced in advanced lung cancer (33). Thus, high KLRB1 expression is a protective effect in most tumors. In this study, we analyzed the expression of KLRB1 in LUAD and its prognosis by analyzing high-throughput sequencing data using bioinformatics methods. Meanwhile, the relationship between KLRB1 and TIIC was analyzed using algorithms to explore the effect of KLRB1 on the immune microenvironment of lung cancer. Finally, the role and specific mechanism of KLRB1 on the growth and metastasis of LUAD cells were explored by molecular cytology experiments and animal experiments.
The TCGA database was utilized to assess the levels of mRNA expression in both lung cancer and normal tissues in this study. Analysis and validation of our results revealed that KLRB1 expression was significantly reduced in LUAD tissues at both the mRNA and protein levels. This reduction was found to be linked to unfavorable clinical characteristics such as T stage, M stage, race, age, sex, smoking status, pathological stage, and first-line treatment outcome. Additionally, lung cancer patients with low KLRB1 expression have significantly lower OS than those with high KLRB1 expression (P<0.05). KLRB1, T stage, N stage, and M stage. KLRB1 was found to have an impact on OS according to univariate Cox analysis. Furthermore, multifactorial analysis revealed that KLRB1 has the potential to be a prognostic biomarker for LUAD.
Previous studies on various cancers have shown a substantial correlation between CD161, encoded by KLRB1, and immune-related pathways. Specifically, KLRB1 was found to be correlated with lymphoid and nonlymphocytic pathways involved in immunomodulation (34). After extensive research on the impact of KLRB1 on LUAD progression was conducted, GSEA was performed and revealed strong correlations between KLRB1 and various crucial immune response pathways, including cytokine and cytokine receptor interactions, antigen processing and presentation, NK cell-mediated cytotoxicity, the T-cell receptor signaling pathway, the B-cell receptor signaling pathway, leukocyte transendothelial migration, and the JAK-STAT signaling pathway. These findings further support the importance of KLRB1 in immunomodulation in LUAD.
Tumor cells grow in a complex microenvironment composed of cancer cells, immune cells and stromal cells, and the relationship between tumor cells and the immune system is determined by a complex network of intercellular interactions. Over the past decade, the TME has become a key factor influencing tumor development, treatment resistance and prognosis (35-37). TIICs have the ability to control cancer progression and may serve as potential indicators of disease prognosis (38). In addition, TIICs have been shown to predict the response to neoadjuvant chemotherapy and immune checkpoint inhibition (ICI) therapy (39). Additionally, the analysis revealed a significant association between KLRB1 and cytokine interactions as well as immune cell response regulatory pathways. The correlation between the expression of KLRB1 and the levels of infiltrating immune cells was assessed. We found that KLRB1 expression was most strongly associated with T cells, cytotoxic cells, B cells, Th1 cells and CD8+ T cells. In non-small cell lung cancer, several studies have shown that tumor-infiltrating T cells are associated with a better prognosis (40-42). It is widely recognized that CD8+ T cells play a crucial role in the cellular immune system and are essential for effective cell-mediated antitumor immune responses (43,44). In lung cancer, CD8+ T cells are an independent prognostic factor and are associated with a better prognosis (41). Reduced survival in LUAD patients is associated with a decrease in the number of B cells among tumor-infiltrating lymphocytes (45). Th1 cells produce the cytokines IL-2, IL-12 and IFN-γ, which exert antitumor effects in humans (46). KLRB1 may play a role in the development and outcome of LUAD by controlling the infiltration of immune cells that have the ability to combat tumors. Accordingly, we constructed immune gene signatures for LUAD based on KLRB1-associated immunomodulators. Thus, the differential expression of KLRB1-associated immunomodulators can distinguish between risk groups.
Furthermore, we performed in vitro experiments to explore the potential biological roles of KLRB1 in LUAD. In vitro, we found that KLRB1 was downregulated in LUAD tissues. Knocking down KLRB1 in A549 cells had a positive influence on the proliferative, migratory and invasive properties of LUAD cells. An analysis of the aforementioned results suggested that KLRB1 plays an important role in controlling disease progression in LUAD patients.
While this study has some limitations, it expands our knowledge regarding the connection between KLRB1 and LUAD. First, this study’s focus was limited to a single dataset, potentially leading to bias stemming from biased selection. Second, following this investigation of the relationship between KLRB1 and immune infiltration in LUAD patients, additional experiments are needed to verify the role of KLRB1 in influencing the TME of LUAD.
Conclusions
In conclusion, low KLRB1 expression is an independent adverse prognostic factor for LUAD and is closely associated with aggressive clinical features and unfavorable immune infiltration. The prognostic signature based on KLRB1-related immunomodulators is an independent predictor of OS in patients with LUAD. In terms of the potential clinical impact, KLRB1 may inhibit the progression of LUAD by suppressing cell proliferation and differentiation via the MAPK signaling pathway.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Reporting Checklist: The authors have completed the MDAR and TRIPOD reporting checklists. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-8/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-8/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-8/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-8/coif). All authors report grants from the National Natural Science Foundation of China (Nos. 81972173 and 82273371) and the Science and Technology Commission of Shanghai Municipality (No. 22140901400). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study of non-small cell lung cancer tissue samples was in accordance with the Ethics Committee of Huashan Hospital, Fudan University (2022 Pro Review No. 634). Written informed consent was obtained from all participating patients for publication.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Ma J, Ward EM, Smith R, et al. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 2013;119:1381-5. [Crossref] [PubMed]
- Pleshkan VV, Zinov'eva MV, Vinogradova TV, et al. Transcription of the KLRB1 gene is suppressed in human cancer tissues. Mol Gen Mikrobiol Virusol 2007;3-7.
- Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol 1994;153:2417-28.
- Rosen DB, Bettadapura J, Alsharifi M, et al. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol 2005;175:7796-9. [Crossref] [PubMed]
- Exley M, Porcelli S, Furman M, et al. CD161 (NKR-P1A) costimulation of CD1d-dependent activation of human T cells expressing invariant V alpha 24 J alpha Q T cell receptor alpha chains. J Exp Med 1998;188:867-76. [Crossref] [PubMed]
- Aldemir H, Prod'homme V, Dumaurier MJ, et al. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol 2005;175:7791-5. [Crossref] [PubMed]
- Fergusson JR, Fleming VM, Klenerman P. CD161-expressing human T cells. Front Immunol 2011;2:36. [Crossref] [PubMed]
- Germain C, Meier A, Jensen T, et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-γ contributes to modulate immune responses. J Biol Chem 2011;286:37964-75. [Crossref] [PubMed]
- Braud VM, Biton J, Becht E, et al. Expression of LLT1 and its receptor CD161 in lung cancer is associated with better clinical outcome. Oncoimmunology 2018;7:e1423184. [Crossref] [PubMed]
- Mathewson ND, Ashenberg O, Tirosh I, et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 2021;184:1281-1298.e26. [Crossref] [PubMed]
- Li Z, Zheng B, Qiu X, et al. The identification and functional analysis of CD8+PD-1+CD161+ T cells in hepatocellular carcinoma. NPJ Precis Oncol 2020;4:28. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Meijuan C, Meng X, Fang L, et al. Synaptotagmin-like protein 1 is a potential diagnostic and prognostic biomarker in endometrial cancer based on bioinformatics and experiments. J Ovarian Res 2023;16:16. [Crossref] [PubMed]
- Zhao W, Lin J, Cheng S, et al. Comprehensive analysis of COMMD10 as a novel prognostic biomarker for gastric cancer. PeerJ 2023;11:e14645. [Crossref] [PubMed]
- Xie J, Lin Y, Li Y, et al. lncRNA TRHDE-AS1 Correlated with Genomic Landscape and Clinical Outcome in Glioma. Genes (Basel) 2023;14:1052. [Crossref] [PubMed]
- Zhang X, Zhang X, Jiang D, et al. INHA acts as a novel and potential biomarker in lung adenocarcinoma and shapes the immune-suppressive tumor microenvironment. Transl Oncol 2023;33:101679. [Crossref] [PubMed]
- Yang X, Su X, Wang Z, et al. ULBP2 is a biomarker related to prognosis and immunity in colon cancer. Mol Cell Biochem 2023;478:2207-19. [Crossref] [PubMed]
- Triozzi PL, Stirling ER, Song Q, et al. Circulating Immune Bioenergetic, Metabolic, and Genetic Signatures Predict Melanoma Patients' Response to Anti-PD-1 Immune Checkpoint Blockade. Clin Cancer Res 2022;28:1192-202. [Crossref] [PubMed]
- Yuan Q, Deng D, Pan C, et al. Integration of transcriptomics, proteomics, and metabolomics data to reveal HER2-associated metabolic heterogeneity in gastric cancer with response to immunotherapy and neoadjuvant chemotherapy. Front Immunol 2022;13:951137. [Crossref] [PubMed]
- Xiao Y, Bi M, Guo H, et al. Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis. EBioMedicine 2022;79:104001. [Crossref] [PubMed]
- Liu XN, Cui DN, Li YF, et al. Multiple "Omics" data-based biomarker screening for hepatocellular carcinoma diagnosis. World J Gastroenterol 2019;25:4199-212. [Crossref] [PubMed]
- He JR, Li D, Zhang QX, et al. Inhibiting KLRB1 expression is associated with impairing cancer immunity and leading to cancer progression and poor prognosis in breast invasive carcinoma patients. Aging (Albany NY) 2023;15:13265-86. [Crossref] [PubMed]
- Xu N, Meng X, Chu H, et al. The prognostic significance of KLRB1 and its further association with immune cells in breast cancer. PeerJ 2023;11:e15654. [Crossref] [PubMed]
- Liang C, Chen Y, Chen S, et al. KLRB1 is a novel prognostic biomarker in endometrial cancer and is associated with immune infiltration. Transl Cancer Res 2023;12:3641-52. [Crossref] [PubMed]
- Wei Y, Xu T, Li C, et al. CD161 Characterizes an Inflamed Subset of Cytotoxic T Lymphocytes Associated with Prolonged Survival in Human Papillomavirus-Driven Oropharyngeal Cancer. Cancer Immunol Res 2023;11:306-19. [Crossref] [PubMed]
- Yin Y, Luo Y, He K. Landscape of immunocytes infiltration and prognostic immune-related genes in hepatocellular carcinoma. Asian J Surg 2023;46:4251-60. [Crossref] [PubMed]
- Lu H, Qian J, Cheng L, et al. Single-cell RNA-sequencing uncovers the dynamic changes of tumour immune microenvironment in advanced lung adenocarcinoma. BMJ Open Respir Res 2023;10:e001878. [Crossref] [PubMed]
- Zhou X, Du J, Liu C, et al. A Pan-Cancer Analysis of CD161, a Potential New Immune Checkpoint. Front Immunol 2021;12:688215. [Crossref] [PubMed]
- Chen F, Zhuang X, Lin L, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med 2015;13:45. [Crossref] [PubMed]
- Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett 2017;387:61-8. [Crossref] [PubMed]
- Liu Y, Guo J, Huang L. Modulation of tumor microenvironment for immunotherapy: focus on nanomaterial-based strategies. Theranostics 2020;10:3099-117. [Crossref] [PubMed]
- Zhang J, Wang F, Xu J, et al. Micro ribonucleic acid-93 promotes oncogenesis of cervical cancer by targeting RAB11 family interacting protein 1. J Obstet Gynaecol Res 2016;42:1168-79. [Crossref] [PubMed]
- Denkert C, Loibl S, Noske A, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 2010;28:105-13. [Crossref] [PubMed]
- Schalper KA, Brown J, Carvajal-Hausdorf D, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 2015;107:dju435. [Crossref] [PubMed]
- Donnem T, Hald SM, Paulsen EE, et al. Stromal CD8+ T-cell Density—A Promising Supplement to TNM Staging in Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:2635-43. [Crossref] [PubMed]
- Geng Y, Shao Y, He W, et al. Prognostic Role of Tumor-Infiltrating Lymphocytes in Lung Cancer: a Meta-Analysis. Cell Physiol Biochem 2015;37:1560-71. [Crossref] [PubMed]
- Barry M, Bleackley RC. Cytotoxic T lymphocytes: all roads lead to death. Nat Rev Immunol 2002;2:401-9. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Lee HE, Luo L, Kroneman T, et al. Increased Plasma Cells and Decreased B-cells in Tumor Infiltrating Lymphocytes are Associated with Worse Survival in Lung Adenocarcinomas. J Clin Cell Immunol 2020;11:584.
- Becker Y. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy--a review. Virus Genes 2006;33:235-52. [Crossref] [PubMed]


