
Differences in T cell-associated serum markers between ischemic cardiomyopathy and dilated cardiomyopathy
Highlight box
Key findings
• Chemokine (C-C motif) ligand 21 (CCL21), interleukin (IL)-1β, lymphocyte-activation gene 3 (LAG3), and vascular cell adhesion molecule-1 (VCAM-1) were found to be able to distinguish between ischemic cardiomyopathy (ICM) and dilated cardiomyopathy (DCM).
What is known and what is new?
• Differentiating between ICM and DCM is critical for clinical management, and pathogenesis-based serological indicators are the future of rapid differentiation between ICM and DCM.
• Four serum markers were identified to distinguish between ICM and DCM.
What is the implication, and what should change now?
• We can quickly distinguish between ICM and DCM by measuring CCL21, IL-1β, LAG3, and VCAM-1 in serum without the need for cumbersome imaging tests. And this can help doctors quickly determine treatment protocols in an emergency clinic.
Introduction
Heart failure (HF) is the terminal stage of the progression of various cardiovascular diseases, and is one of the main direct causes of death in patients with cardiovascular diseases, with reduced exercise tolerance, respiratory weakness and edema, and the mortality rate of patients within 5 years is as high as 60% (1,2). Ischemic cardiomyopathy (ICM) and dilated cardiomyopathy (DCM) are the two most important diseases that cause HF, and although the clinical manifestations of patients with these diseases are basically the same, the clinical treatment protocols are completely different due to their different pathogeneses (3,4). Therefore, differentiation of which cardiomyopathy is causing HF would be necessary but may not be possible in the emergency department.
Currently, coronary angiography (CAG) is the gold standard for differentiating between ICM and DCM (5,6). However, as an invasive test, CAG not only has certain risks, but is also very expensive, and many grassroots medical institutions are limited by equipment and personnel constraints to carry out CAG testing (7,8). Serological indicators are important auxiliary indicators for clinical differential diagnosis of various diseases, which have the advantage of rapidity and low equipment and personnel requirements. However, there are currently few serologic markers that can be used to distinguish between ICM and DCM, mainly due to the current lack of understanding of the pathogenesis of DCM. With the development of high-throughput sequencing, multiple pathological mechanisms related to the pathogenesis of DCM have been discovered, including infectious or non-infectious inflammation, autoimmune diseases, endocrine and metabolic disorders, poisoning, and genetic diseases (9,10). In addition, a number of new biomarkers for improving cardiac function, such as sodium/glucose cotransporter 2 (SGLT2), have also been screened (11,12). The pathogenesis of ICM is relatively clear, that is, myocardial injury caused by long-term ischemia and hypoxia of the myocardium caused by coronary arteriosclerosis (13). Therefore, pathogenesis-based serological indicators are the future of rapid differentiation between ICM and DCM.
In this study, we used transcriptome sequencing data from heart tissue in public databases (GSE116250, a set of RNA-sequencing data on cardiac tissue from patients with DCM and ICM) to analyze the differences in pathogenesis between ICM and DCM, and then looked for secreted proteins from the hub genes involved in the pathogenesis. Potential serological markers to distinguish between ICM and DCM were identified by detecting and comparing these differentially expressed genetically encoded secreted proteins in serum. We present this article in accordance with the STARD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-901/rc).
Methods
Patients and ethics statement
From January 2022 to December 2023 in Department of Cardiovascular Medicine, The First Affiliated Hospital of Bengbu Medical University, we enrolled 97 patients hospitalized for HF which was diagnosed according to 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic HF (14), including 48 patients with DCM and 49 patients with ICM. At the same time, we also enrolled 25 age- and sex-matched healthy volunteers. Demographic and clinical characteristics of all participants are shown in Table 1. The inclusion and exclusion criteria for patients with HF were as follows. Inclusion criteria: (I) patients with ICM are diagnosed with stenosis of at least 1 coronary artery by CAG; (II) patients with DCM are diagnosed by CAG without coronary artery stenosis; (III) complete clinical data. Exclusion criteria: (I) comorbid with other types of heart disease; (II) combined with severe liver and kidney insufficiency, hoof tissue disease, malignant tumors, thyroid diseases, and severe infectious diseases; (III) record of invasive treatment within half a year; (IV) infectious diseases, drug addiction, or alcohol addiction; (V) pregnant or lactating females; (VI) diseases of the immune system or blood system. This study protocol was reviewed and annotated by the Ethics Committee of The First Affiliated Hospital of Bengbu Medical University (approval number [2023]ky039), and the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all individual participants.
Table 1
| Variables | Healthy (N=25) | DCM (N=48) | ICM (N=49) | P value |
|---|---|---|---|---|
| Age (years) | 55.32±6.71 | 56.24±6.35 | 56.27±7.03 | 0.42 |
| Male | 15 (60.00) | 34 (70.83) | 36 (73.46) | 0.72 |
| BMI (kg/m2) | 22.12±3.41 | 22.41±2.51 | 21.99±4.02 | 0.36 |
| NYHA grade, n (%) | 0.50 | |||
| II | – | 18 (37.50) | 19 (38.78) | |
| III | – | 24 (50.00) | 20 (40.82) | |
| IV | – | 6 (12.50) | 10 (20.41) |
Data are presented as mean ± standard deviation or n (%). P value indicates difference between DCM group and ICM group. BMI, body mass index; NYHA, New York Heart Association; DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy.
Cardiac transcriptome sequencing data analysis
There were 14 non-failing donors (NFD), 37 DCM patients, and 13 ICM patients who had contributed their left ventricle for transcriptome sequencing, and RNA-sequencing data had been uploaded to the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) by Sweet et al. and Yamaguchi et al. (15,16). We used the GEO2R tool on the National Center for Biotechnology Information (NCBI) platform to identify differentially expressed genes (DEGs) among ICM versus NFD, DCM versus NFD, and DCM versus ICM, and the Bioinformatics online tool (http://www.bioinformatics.com.cn) to visualize DEGs as volcano maps. At the same time, we functionally enriched DEGs in the Gene Ontology (GO) database (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/) and visualized them using the Bioinformatics online tool. In addition, we extracted the data of gene expression, and visualization was performed using the heatmaps package in R software (https://bioconductor.org/packages/release/bioc/html/heatmaps.html). Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithm was used to evaluate immune cells infiltration in the heart tissues (LM22 signature, 1,000 permutations) (17,18).
Screening of hub-genes analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online database (https://cn.string-db.org/) was used to analyze protein-molecule interactions, namely protein-protein interaction (PPI) networks. And then we exported score results of molecular interconnection to screen top 10 hub-genes.
Detection of serum CCL21, IL-1β, LAG3, and VCAM-1
In the early morning fasting condition, 5–10 mL of peripheral blood was collected from all participants and centrifuged (room temperature and 1,000 ×g) to obtain serum. The serum levels of chemokine (C-C motif) ligand 21 (CCL21), interleukin (IL)-1β, lymphocyte-activation gene 3 (LAG3), and vascular cell adhesion molecule-1 (VCAM-1) were determined using a human exodus 2 (CCL21) enzyme-linked immunosorbent assay (ELISA) kit (ab193759, Abcam, Cambridge, UK), human IL-1 beta ELISA kit (ab214025, Abcam), human LAG-3 ELISA kit (ab215061, Abcam), and human VCAM-1 ELISA kit (ab223591, Abcam), respectively.
Statistical analysis
Measurement data in this study were expressed as (mean ± standard deviation) and counting data were shown as n (%). The software SPSS 20.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The chi-square test was used to compare the differences in counting data between the two groups, and the Student’s t-test was used to compare the differences in quantitative data between the two groups. One-way analysis of variance (ANOVA) was used for continuous data comparison among the three groups. Receiver operating characteristic (ROC) curve was used to evaluate the diagnostic efficacy of serum CCL21, IL-1β, LAG3, and VCAM-1 in patients with ICM. A P value <0.05 indicated a significant difference.
Results
Identification and enrichment analysis of DEGs versus NFD
Transcriptome sequencing was performed on the left ventricles of 14 NFD, 37 DCM, and 13 ICM. Firstly, we compared transcriptome data between ICM and NFD groups to identify DEGs in ICM patients, and there were 1,263 DEGs that met the screening requirements [|log2 fold change (FC)| >1 and adjusted P<0.05], comprising 418 DEGs of down-regulation and 845 DEGs of up-regulation (Figure 1A). Functional enrichment analysis of DEGs in the GO database showed that there were altogether 1,986 features enriched, including T cell activation, leukocyte cell-cell adhesion, cytokine activity, cytokine receptor binding, immune receptor activity, receptor ligand activity, signaling receptor activator activity, and so on (Figure 1B and available online: https://cdn.amegroups.cn/static/public/jtd-24-901-1.xlsx). Besides, the results of enrichment analysis of DEGs in the KEGG database showed a total of 74 pathways, including cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway, cell adhesion molecules, Th17 cell differentiation, and Th1 and Th2 cell differentiation, among others (Figure 1C and available online: https://cdn.amegroups.cn/static/public/jtd-24-901-1.xlsx).
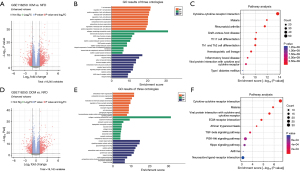
In addition, there were 956 DEGs between DCM versus NFD that met the screening requirements (|log2 FC| >1 and adjusted P<0.05), comprising 341 DEGs of down-regulation and 615 DEGs of up-regulation (Figure 1D). Functional enrichment analysis of DEGs in the GO database showed that altogether 1,328 features were enriched, including extracellular matrix organization, extracellular structure organization, modulation of chemical synaptic transmission, regulation of trans-synaptic signaling and cellular calcium ion homeostasis, and so on (Figure 1E and available online: https://cdn.amegroups.cn/static/public/jtd-24-901-2.xlsx). Besides, the results of enrichment analysis of DEGs in the KEGG database showed a total of 36 pathways, including cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, ECM-receptor interaction, African trypanosomiasis, and transforming growth factor (TGF)-beta signaling pathway (Figure 1F and available online: https://cdn.amegroups.cn/static/public/jtd-24-901-2.xlsx).
Screening DEGs to identify ICM
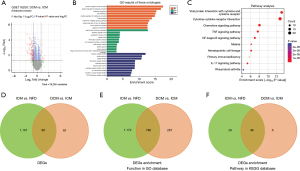
First of all, there were 158 DEGs between DCM and ICM that met the screening requirements (|log2 FC| >1 and adjusted P<0.05), comprising 147 DEGs of down-regulation and 11 DEGs of up-regulation (Figure 2A). Functional enrichment analysis of DEGs in the GO database showed that altogether 1,053 features were enriched, including positive regulation of cell activation, positive regulation of leukocyte activation, leukocyte chemotaxis, cell chemotaxis, positive regulation of lymphocyte activation, T cell activation, and so on (Figure 2B and Table S1). Moreover, the results of enrichment analysis of DEGs in the KEGG database showed a total of 52 pathways, including cytokine-cytokine receptor interaction, chemokine signaling pathway, TNF signaling pathway, PI3K-Akt signaling pathway, IL-17 signaling pathway, Th17 cell differentiation, and Th1 and Th2 cell differentiation (Figure 2C and Table S1). Next, we found that 96 of the 158 DEGs between DCM and ICM were also differentially expressed between ICM and NFD (Figure 2D). Importantly, most of the features (786 of 1,053 features) that were enriched with DEGs between DCM and ICM in the GO database were also present between ICM and NFD (Figure 2E and Figure S1), and the same was revealed by the enrichment analysis results of DEGs in the KEGG database (46 of 52 pathways) (Figure 2F).
Immune infiltration analysis in GSE116250
We analyzed the score of immune cells in GSE116250, and found that there was no significant difference between the NFD group and DCM group (Figure 3A), yet a significant difference in T cells CD8, T cells CD4 memory resting and activated, T cells follicular helper, monocytes, macrophage M1, and mast cells resting between the NFD group and ICM group (Figure 3B), and a significant difference in T cells CD8, T cells CD4 memory resting and activated, T cells follicular helper, NK cells resting, macrophage M1, and macrophage M2 (Figure 3C). Importantly, there was a significant difference in T cells CD8, T cells CD4 memory resting and activated, T cells follicular helper, and macrophage M1 between ICM and NFD or DCM (Figure 3D).
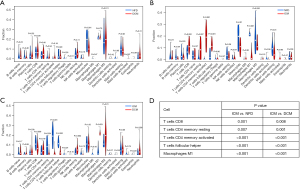
Hub genes associated with T cells
Compared with the ICM group, there were 75 DEGs related to T cells in the NFD group, 25 DEGs related to T cells in the DCM group, and 17 DEGs related to T cells in the NFD and DCM groups (Figure 4A). Analysis of the expression of these 17 DEG-related T cells showed that the expression of all of them in the ICM group were higher than they were in the NFD group and DCM group (Figure 4B). At last, to screen hub genes, we created a PPI network with these 17 DEGs related T cells using the STRING database (Figure 4C and Table S2), and found that two genes were free outside the PPI network (PNP and EGR3), and the top ten genes with the highest link scores were CCL21, IL-1B, LAG3, VCAM-1, CD8A, CD5, IL7R, LCK, TNFRSF4, and ITK.

Serum levels of protein related to T cells
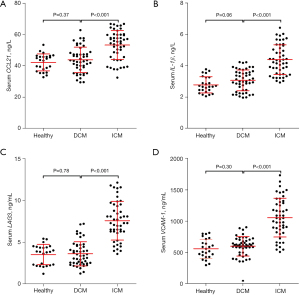
Among the top ten hub genes related to T cells, there were four genes encoding proteins that could be secreted into serum, namely CCL21, IL-1β, LAG3, and VCAM-1. Therefore, we collected 25 healthy controls, 48 patients with DCM, and 49 patients with ICM to determine the serum levels of CCL21, IL-1β, LAG3, and VCAM-1. The results showed that there was no significant difference in the serum levels of CCL21, IL-1β, LAG3, and VCAM-1 between healthy people and DCM patients, whereas the serum levels of CCL21, IL-1β, LAG3, and VCAM-1 in ICM patients were all significantly higher than these in DCM patients (Figure 5).
Diagnostic efficacy of T cell-associated serum markers for ICM
The ROC curve was used to distinguish between ICM and DCM patients, and we found that the area under the curve of the ROC curve (AUC) for serum CCL21, IL-1β, LAG3, and VCAM-1 in 48 patients with DCM and 49 patients with ICM was 0.775, 0.868, 0.934, and 0.903, respectively. At the optimal cut-off value of the ROC curve, the sensitivity of serum CCL21, IL-1β, LAG3, and VCAM-1 in the diagnosis of ICM patients were 73.47%, 71.43%, 79.17%, and 81.63%, and the specificity were 77.08%, 85.42%, 91.67%, and 93.75%, respectively (Figure 6 and Table 2).
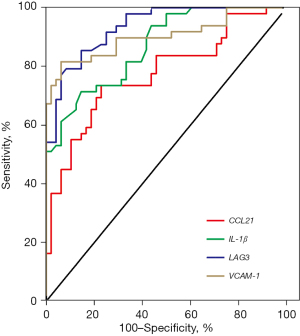
Table 2
| Index | AUC (95% CI) | Cutoff value | Specificity (%) | Sensitivity (%) |
|---|---|---|---|---|
| CCL2 | 0.775 (0.681−0.868) | 48.20 ng/L | 77.08 | 73.47 |
| IL-1β | 0.868 (0.800−0.936) | 3.835 ng/L | 85.42 | 71.43 |
| LAG3 | 0.934 (0.890−0.979) | 6.075 ng/mL | 91.67 | 79.17 |
| VCAM-1 | 0.903 (0.839−0.967) | 794.9 ng/mL | 93.75 | 81.63 |
ICM, ischemic cardiomyopathy; DCM, dilated cardiomyopathy; CCL21, chemokine (C-C motif) ligand 21; IL-1β, interleukin-1β; LAG3, lymphocyte-activation gene 3; VCAM-1, vascular cell adhesion molecule-1; AUC, area under the curve; CI, confidence interval.
Discussion
ICM is a heart disease mainly caused by narrowing or blockage of the coronary arteries, whereas DCM is caused by an enlarged and deformed heart. Although patients with ICM and DCM have similar clinical presentations, their pathogenesis and pathology are completely different. In terms of pathogenesis, ICM is caused by the lack of oxygen to the heart muscle cells due to insufficient blood supply to the coronary arteries, which causes a series of pathological changes (19,20), whereas DCM is caused by the breakdown of elastic fibers in the heart wall, causing the heart to gradually continuously expand (9,21). In addition, myocardial ischemia is a direct cause of ICM, and there are many causes of DCM (13), which is the main reason why ICM and DCM are difficult to distinguish (7,8). Herein, we found that compared with NFD heart tissue, the DEGs function of ICM patients was mainly enriched in T cell activation and differentiation, cytokines, and cell-cell adhesion, whereas the DEGs function of DCM patients was mainly enriched in extracellular matrix organization, extracellular structure organization, modulation of chemical synaptic transmission, and so on. Therefore, these results indicated that there is a difference between cardiac histopathological changes in patients with ICM and DCM, and that it is feasible to quickly distinguish between ICM and DCM using serological indicators based on pathogenesis.
In order to explore the differences in the pathogenesis and pathological changes of ICM and DCM, we further compared the transcriptome sequencing data of cardiac tissues of ICM and DCM patients, and found that many DEGs in the heart tissues of DCM patients were also differentially expressed in NFD compared with ICM. Analysis of these intersecting DEGs and the functions enriched by these DEGs revealed that immune cells (especially T cells and leukocytes), chemokines, cytokines, and inflammation are the most common characteristics of these DEGs. In recent years, mounting studies have confirmed that the immune microenvironment is essential for the occurrence and progression of ICM (22-24); Bansal et al. found that pro-inflammatory T lymphocytes were crucial for cardiac remodeling in patients with ICM (23), and study by Xia et al. suggested that the unique regulatory T lymphocyte population of cardiac tissue infiltration had a protective effect on the heart of patients with myocardial infarction (24). As previously reported, inflammation is a risk factor for the development of cardiovascular disease (25), and some biomarkers associated with inflammation can be used to assess the clinical prognosis of patients with HF (26). At the same time, based on the results of the above transcriptome sequencing data, we shifted our focus to the immune microenvironment of cardiac tissue in patients with ICM and DCM. The results of immune cell scoring showed that although there was no difference between NFD and DCM, they were significantly different from ICM patients in CD8 T cells CD4 T cells memory resting and activated, T cells follicular helper, and M1 macrophage, suggesting that the activation/differentiation of T cells and macrophage polarization in the immune microenvironment of cardiac tissue may be key to differentiating ICM from DCM. Since macrophage polarization is non-specific, and that acute or chronic inflammatory responses are considered a common factor in the pathogenesis of ICM and DCM, we will focus on studying T cell-related genes in future work. Returning to the functional enrichment results of differential DEGs, we selected the gene set of DEGs associated with T cells for further analysis. Finally, four T cell-related genes encoding secreted proteins were identified, namely CCL21, IL-1β, LAG3, and VCAM-1.
CCL21 is a chemokine that acts on T cells but has no chemotaxis activity against B cells and macrophages (27,28). At the same time, CCL21 promotes T cell migration by binding to the PSGL-1 glycoprotein on T cells. IL-1, also known as lymphocyte-stimulating factor, mainly exists in two forms: IL-1α and IL-1β, and their main function is to promote the activation, proliferation and differentiation of thymic T cells (29). LAG-3 protein is an inhibitory receptor on antigen-activated T cells, which, when bound to FGL1, transmits inhibitory signals that negatively regulate the proliferation, activation, effector function, and homeostasis of CD8+ and CD4+ T cells (30). VCAM-1 is a ligand for very late antigen 4 (VLA-4) present on leukocytes, and its mediated cell-to-cell adhesion is essential for T cell activation and leukocyte recruitment to the site of inflammation (31).
To verify the value of the 4 T cell-associated genes identified above in differentiating between ICM and DCM, we recruited NFDs, DCM patients, and ICM patients to measure their serum levels. We found that the serum levels of CCL21, IL-1β, LAG3, and VCAM-1 in ICM patients were significantly higher than those in DCM patients, and the ROC curves showed that serum levels of CCL21, IL-1β, LAG3, and VCAM-1 had good diagnostic efficacy in the diagnosis of ICM. All in all, we identified four T cell-associated serum markers that could be used to distinguish between ICM and DCM.
Based on cardiac transcriptome data, we identified four differentially expressed genes in ICM patients and DCM patients, which encode secreted proteins. We further identified these four protein levels secreted into the serum, i.e., serum CCL21, IL-1β, LAG3, and VCAM-1, that can be used to distinguish between ICM and DCM. However, it should be noted that the smaller sample size is a limitation of this study, and the inclusion of more patients will be more meaningful for the validation of the results of this study. All in all, we have identified 4 T cell-associated serum markers, CCL21, IL-1β, LAG3, and VCAM-1, as potential diagnostic serum markers that differentiate ICM from DCM.
Conclusions
All in all, results in the present study indicated that four T cell-associated serum markers had been identified as potential diagnostic serum markers that differentiate ICM from DCM, that is, CCL21, IL-1β, LAG3, and VCAM-1.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-901/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-901/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-901/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-901/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study protocol was reviewed and annotated by the Ethics Committee of The First Affiliated Hospital of Bengbu Medical University (approval number [2023]ky039). Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Emmons-Bell S, Johnson C, Roth G. Prevalence, incidence and survival of heart failure: a systematic review. Heart 2022;108:1351-60. [Crossref] [PubMed]
- Savarese G, Becher PM, Lund LH, et al. Global burden of heart failure: a comprehensive and updated review of epidemiology. Cardiovasc Res 2023;118:3272-87. [Crossref] [PubMed]
- Liu M, Zhai L, Yang Z, et al. Integrative Proteomic Analysis Reveals the Cytoskeleton Regulation and Mitophagy Difference Between Ischemic Cardiomyopathy and Dilated Cardiomyopathy. Mol Cell Proteomics 2023;22:100667. [Crossref] [PubMed]
- Shao X, Zhang X, Yang L, et al. Integrated analysis of mRNA and microRNA expression profiles reveals differential transcriptome signature in ischaemic and dilated cardiomyopathy induced heart failure. Epigenetics 2021;16:917-32. [Crossref] [PubMed]
- Wintrich J, Berger AK, Bewarder Y, et al. Update on diagnostics and treatment of heart failure. Herz 2022;47:340-53. [Crossref] [PubMed]
- Heidenreich PA, Bozkurt B, Aguilar D, et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol 2022;79:1757-80. [Crossref] [PubMed]
- Ngam PI, Ong CC, Chai P, et al. Computed tomography coronary angiography - past, present and future. Singapore Med J 2020;61:109-15. [Crossref] [PubMed]
- Graby J, Khavandi A, Thompson D, et al. CT coronary angiography-guided cardiovascular risk screening in asymptomatic patients: is it time? Clin Radiol 2021;76:801-11. [Crossref] [PubMed]
- Heymans S, Lakdawala NK, Tschöpe C, et al. Dilated cardiomyopathy: causes, mechanisms, and current and future treatment approaches. Lancet 2023;402:998-1011. [Crossref] [PubMed]
- Reichart D, Magnussen C, Zeller T, et al. Dilated cardiomyopathy: from epidemiologic to genetic phenotypes: A translational review of current literature. J Intern Med 2019;286:362-72. [Crossref] [PubMed]
- Marfella R, D'Onofrio N, Trotta MC, et al. Sodium/glucose cotransporter 2 (SGLT2) inhibitors improve cardiac function by reducing JunD expression in human diabetic hearts. Metabolism 2022;127:154936. [Crossref] [PubMed]
- Marfella R, Scisciola L, D'Onofrio N, et al. Sodium-glucose cotransporter-2 (SGLT2) expression in diabetic and non-diabetic failing human cardiomyocytes. Pharmacol Res 2022;184:106448. [Crossref] [PubMed]
- Pastena P, Frye JT, Ho C, et al. Ischemic cardiomyopathy: epidemiology, pathophysiology, outcomes, and therapeutic options. Heart Fail Rev 2024;29:287-99. [Crossref] [PubMed]
- McDonagh TA, Metra M, Adamo M, et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J 2021;42:3599-726. [Crossref] [PubMed]
- Sweet ME, Cocciolo A, Slavov D, et al. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genomics 2018;19:812. [Crossref] [PubMed]
- Yamaguchi T, Sumida TS, Nomura S, et al. Cardiac dopamine D1 receptor triggers ventricular arrhythmia in chronic heart failure. Nat Commun 2020;11:4364. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Craven KE, Gökmen-Polar Y, Badve SS. CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer. Sci Rep 2021;11:4691. [Crossref] [PubMed]
- Del Buono MG, Moroni F, Montone RA, et al. Ischemic Cardiomyopathy and Heart Failure After Acute Myocardial Infarction. Curr Cardiol Rep 2022;24:1505-15. [Crossref] [PubMed]
- Dang H, Ye Y, Zhao X, et al. Identification of candidate genes in ischemic cardiomyopathy by gene expression omnibus database. BMC Cardiovasc Disord 2020;20:320.
- Ampong I. Metabolic and Metabolomics Insights into Dilated Cardiomyopathy. Ann Nutr Metab 2022;78:147-55. [Crossref] [PubMed]
- Sun K, Li YY, Jin J. A double-edged sword of immuno-microenvironment in cardiac homeostasis and injury repair. Signal Transduct Target Ther 2021;6:79. [Crossref] [PubMed]
- Bansal SS, Ismahil MA, Goel M, et al. Dysfunctional and Proinflammatory Regulatory T-Lymphocytes Are Essential for Adverse Cardiac Remodeling in Ischemic Cardiomyopathy. Circulation 2019;139:206-21. [Crossref] [PubMed]
- Xia N, Lu Y, Gu M, et al. A Unique Population of Regulatory T Cells in Heart Potentiates Cardiac Protection From Myocardial Infarction. Circulation 2020;142:1956-73. [Crossref] [PubMed]
- Sardu C, Paolisso G, Marfella R. Inflammatory Related Cardiovascular Diseases: From Molecular Mechanisms to Therapeutic Targets. Curr Pharm Des 2020;26:2565-73. [Crossref] [PubMed]
- Sardu C, Marfella R, Santamaria M, et al. Stretch, Injury and Inflammation Markers Evaluation to Predict Clinical Outcomes After Implantable Cardioverter Defibrillator Therapy in Heart Failure Patients With Metabolic Syndrome. Front Physiol 2018;9:758. [Crossref] [PubMed]
- Zhang N, Liu X, Qin J, et al. LIGHT/TNFSF14 promotes CAR-T cell trafficking and cytotoxicity through reversing immunosuppressive tumor microenvironment. Mol Ther 2023;31:2575-90. [Crossref] [PubMed]
- Loef EJ, Sheppard HM, Birch NP, et al. Plasminogen and plasmin can bind to human T cells and generate truncated CCL21 that increases dendritic cell chemotactic responses. J Biol Chem 2022;298:102112. [Crossref] [PubMed]
- Sylvester PA, Corbett JA, Tarakanova VL. T cell-extrinsic IL-1 signaling controls long-term gammaherpesvirus infection by suppressing viral reactivation. Virology 2022;576:134-40. [Crossref] [PubMed]
- Maruhashi T, Sugiura D, Okazaki IM, et al. Binding of LAG-3 to stable peptide-MHC class II limits T cell function and suppresses autoimmunity and anti-cancer immunity. Immunity 2022;55:912-924.e8. [Crossref] [PubMed]
- Riegler J, Gill H, Ogasawara A, et al. VCAM-1 Density and Tumor Perfusion Predict T-cell Infiltration and Treatment Response in Preclinical Models. Neoplasia 2019;21:1036-50. [Crossref] [PubMed]


