
Identification of a prognostic gene signature in patients with cisplatin resistant squamous cell lung cancer
Highlight box
Key findings
• We established an integrated prognostic signature for patients with lung squamous cell carcinoma (LUSC) and cisplatin resistance. We identified several small-molecule drugs including sulforaphane and parthenolide for improving the survival of patients with LUSC.
What is known and what is new?
• The respective effects of messenger RNA (mRNA), microRNA (miRNA), and alternative splicing (AS) on tumors is widely known. Previous studies have found that sulforaphane and parthenolide exert tumor-suppressive effects.
• In this study, mRNAs, miRNAs, and AS events were integrated, and a prognostic signature was constructed for patients with LUSC and cisplatin resistance. This study further confirmed that sulforaphane and parthenolide could inhibit the growth of LUSC through 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide assay.
What is the implication, and what should change now?
• This study may provide a valuable reference for exploring the underlying mechanisms and therapeutic targets of cisplatin resistance in LUSC. Further research is to explore the role of these key genes in patients with cisplatin resistance in LUSC.
Introduction
Lung cancer ranks third among all new cases of cancer and ranks first among all cancer-related deaths worldwide (1). It was estimated that approximately 238,340 new cases and 127,070 new deaths occurred in the United States in 2023 (1). The most common type of lung cancer is non-small cell lung cancer (NSCLC), which mainly includes lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell cancer based on histology and pathogenesis (2). LUSC represents approximately 30% of all lung cancer cases. Due to the high degree of malignancy and a lack of effective targeted therapeutic drugs, patients with LUSC have a poor prognosis, with a 5-year survival rate of <15% (3,4).
Currently, cisplatin is used as a first-line therapy for several cancers, including NSCLC (5). Cisplatin induces apoptosis of tumor cells through its effects on cell DNA damage where it forms inter- and intra-strand breaks (6,7). Unfortunately, although most patients initially respond to cisplatin treatment, chemotherapy resistance ultimately develops, leading to treatment failure and subsequent relapse (8-11). Therefore, a better understanding of the molecular mechanisms underlying cisplatin resistance in patients with LUSC is critical in devising more effective treatments.
Alternative splicing (AS) refers to the process of generating different messenger RNA (mRNA) splice isoforms through various splicing types from a pre-mRNA, such that the final protein product is expressed differently or with a mutually antagonistic structure and function (12). A study suggested that AS contributes to the complexity of the proteome (13) and that AS events may downregulate the translation of mRNA isoforms by prematurely degrading stop codons (14). A growing body of evidence indicates that aberrant AS of pre-mRNA could contribute to the development and progression of cancers by producing various abnormal proteins, which play a role in tumor cell proliferation, migration, invasion, and other biological processes (15,16).
In recent years, numerous studies have confirmed the significance of AS in the development of various cancers, including breast cancer, ovarian cancer, and lung cancer, among others (17-19). For example, the AS transcript of ceramide synthase 2 (CERS2) can result in cell proliferation and migration of the luminal B breast cancer subtype (17). Moreover, Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) facilitates ovarian cancer metastasis by promoting RNA binding fox-1 homolog 2 (RBFOX2)-mediated AS (18), while Serine and arginine-rich splicing factor 1 (SRSF1)-dependent AS attenuates tumor-inhibiting activity of bridging integrator 1 (BIN1) in NSCLC (19). Additionally, AS is known to mediate chemotherapy resistance in various tumors. For example, long noncoding RNA (lncRNA) colorectal neoplasia differentially expressed (CRNDE) was reported to diminish gastric cancer chemoresistance through the AS of phosphatidylinositol binding clathrin assembly protein (PICALM) which is regulated by serine and arginine rich splicing factor 6 (SRSF6) (20). However, few studies have systematically examined the survival of patients with LUSC with reference to mRNA, microRNA (miRNA), and AS events. The purpose of this study is to explore the roles of mRNA, miRNA, and AS in predicting the prognosis of cisplatin-resistant LUSC patients, identify a combination of prognostic biomarkers, and lay the foundation for finding potential therapeutic targets and exploring the mechanism of cisplatin resistance in LUSC patients.
The Cancer Genome Atlas (TCGA) project provides a wealth of resources to explore the AS patterns of different cancers (21). A variety of molecular expression profiles and clinical information such as drug responses, provide opportunities and challenges for identifying novel molecular signatures and exploring the mechanisms of drug resistance in LUSC. We therefore systematically analyzed genome-wide LUSC-specific mRNA, miRNA, and AS events from TCGA and developed an integrated approach to identify differentially expressed mRNAs, miRNAs, and AS events relevant to the prognosis of patients receiving cisplatin treatment (Figure 1). Overall, this study may provide a feasible strategy for studying cancer drug resistance and identifying LUSC-associated AS events. We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-827/rc).
Methods
Data retrieval
Gene and miRNA expression (RNAseq), and SpliceSeq data for LUSC in addition to the corresponding clinical characteristics, were downloaded from TCGA data portal (https://portal.gdc.cancer.gov/) (21-24) (Table S1). Drug information was included for the analysis of differentially expressed mRNAs, miRNAs, and AS events between cisplatin-sensitive and -resistant samples in LUSC. Overall, data from 70 patients with LUSC including 50 cisplatin-sensitive and 20 cisplatin-resistant patients (Table S2) were selected for the analysis. Patients with complete response and partial response were categorized under the cisplatin-sensitive group, whereas patients with stable disease or progressive disease were categorized in the cisplatin-resistant group. We recruited 6 pairs of matched fresh-frozen lung tumors and adjacent normal tissues from Liaoning Cancer Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Medical Ethics Committee of Liaoning Cancer Hospital (No. 20210826YG) and informed consent was taken from all the patients.
Analysis of differentially expressed genes (DEGs)
The analysis of DEGs, mRNAs and miRNAs, were determined between the cisplatin-sensitive and cisplatin-resistant groups (P<0.05) using the limma package on R 3.5.3 (The R Foundation for Statistical Computing, Vienna, Austria).
Analysis of AS events
SpliceSeq is used to quantify the splicing levels of mRNA in TCGA, which can calculate a percent spliced-in (PSI) value (from 0 to 1) based on the seven types of AS events for each protein-coding gene provided in the Ensemble gene database (25). The PSI value of AS events was calculated and used to analyze the differential AS events between cisplatin-sensitive and cisplatin-resistant groups.
Establishment of prognostic models
Differentially expressed mRNAs, miRNAs, and AS events were incorporated into univariate Cox analyses to identify potential prognosis-associated factors. Least absolute shrinkage and selection operator (LASSO) Cox regression analysis was performed using the glmnet 4.0 in R package version 3.5.3 (https://www.r-project.org) to choose the most significant factors from which predictive models were constructed based on these factors’ corresponding coefficients. Subsequently, the coefficients of these key factors were used to calculate the patients’ risk scores. Median risk scores were then used to divide patients into two groups (26).
Survival analysis
The Kaplan-Meier survival analyses were used to evaluate patients’ prognosis. Receiver operating characteristic (ROC) curves were constructed to assess the sensitivity and specificity of the signature using the survival ROC in R package. The prognostic independence of the signature and clinical characteristics were analyzed through univariate and multivariate Cox regression analyses using the forest plot in R package. Kaplan-Meier plotter online tools were used to perform survival analysis (https://kmplot.com/analysis). To demonstrate the superiority of the predictive performance of this integrated signature, predictive values were compared between this signature and other published signatures (27-29).
Correlation of signature with tumor-infiltrating immune cell characteristics
The stromal and immune scores were downloaded from the Estimation of Stromal and Immune Cells in Malignant Tumor Tissues Using Expression Data (ESTIMATE; (https://bioinformatics.mdanderson.org/estimate). Single-sample gene set enrichment analysis (ssGSEA) using the R package, GSEAbase, was used to evaluate the immune-related characteristics in the different risk subgroups.
Immunohistochemistry
Sections (4 µm) were cut from formalin-fixed, paraffin-embedded lung tumor and paired normal lung tissues. Sections were deparaffinized in two changes of xylene for 10 min each, hydrated in descending concentrations of ethanol, and then immersed in water. Sections were incubated in 3% (v/v) hydrogen peroxide for 15 min to quench endogenous peroxidases. Following this, 1 mM of ethylene diamine tetraacetic acid (EDTA) buffer (pH 8) was used for antigen retrieval under high pressure. Sections were subjected to blocking for 20 min with pre-diluted 10% horse serum and then incubated with primary antibody for 2 h at room temperature in a humidified chamber and polyclonal anti-rabbit (horseradish peroxidase-labeled) secondary antibody (UltraSensitive SP IHC Kit, Maixin-Bio Co., Fuzhou, China) for 30 min. All antibodies were diluted to their final concentrations using phosphate-buffered saline. The primary antibodies used were anti-cortactin (anti-CTTN) (1:50; Proteintech, Chicago, USA) and anti-carboxypeptidase (anti-CPM) (1:50; Abcam, Cambridge, UK). Sections were incubated in diluted (1 drop in 1 mL of antibody diluent) DAB and chromogen (DAB0031, Fuzhou, China) for 2 min. Stained tissue sections were counterstained with hematoxylin, dehydrated through increasing concentrations of ethanol, and cleared with xylene before being mounted with cover slips. The integrated optical density (IOD) was analyzed using Image-Pro Plus 6.0 software (Media Cybernetics Inc., MD, USA).
Screening of potential small-molecule drugs
The CMap database was used to screen for potential drugs that could attenuate cisplatin resistance in patients with LUSC (https://clue.io/). The 3D structure of candidate small-molecule drugs was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The half-maximal inhibitory concentration (IC50) values of the small-molecule drugs in LUSC cell lines were determined via the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/).
Cell culture
The human LUSC cell line, SKMES1, was purchased from the Cell Resource Center of the Shanghai Institute for Biological Sciences. Cells were cultured at 37 °C with 5% CO2 according to standard protocols. Adherent cultures of SKMES1 cells were maintained in RPMI-1640 medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone) and antibiotics (penicillin 100 U/mL, streptomycin 0.1 mg/mL).
3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay
SKMES1 cells were seeded in 96-well plates at a density of 2×103 cells per well and were cultured for 24 h. Each well contained 100 µL of culture medium with a range of concentrations (0.1, 0.3, 1, 3, 10, 30, 100, 300 µM) of sulforaphane or parthenolide. Each concentration of drug was examined in replicate in addition to blank (media only) and vehicle controls. Following incubation at 48 h, 20 µL of 5 mg/mL MTT solution was added to each well and incubated at 37 °C for 3 h, after which time, the culture media was removed. Adherent cells were dissolved in 200 µL of dimethyl sulfoxide (DMSO). The absorbance at 570 nm was measured to evaluate cell viability using multifunctional enzyme marker (infinite 200Pro, Tecan, Switzerland). The growth inhibition rate was calculated according to the following formula: growth inhibition rate (%) = [1 − (test sample − blank control)/(negative control hole − blank control)] × 100%. The IC50 was calculated with SPSS 22.0 software (IBM Corp., Chicago, IL, USA).
Statistical analysis
All statistical analyses were performed using R (R package version 3.5.3 (https://www.r-project.org), SPSS 22.0 software (IBM Corp., Chicago, USA) and GraphPad Prism 7 (GraphPad Software Inc., San Diego, CA, USA) P<0.05 was considered statistically significant.
Results
Identification of DEGs or miRNAs in cisplatin-resistant LUSC
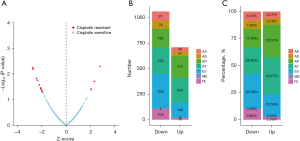
Survival analysis of patients who were cisplatin resistant or sensitive was performed. Resistant patients had poorer overall survival than patients who were sensitive to cisplatin (P<0.001, Figure 2A). Differentially expressed mRNAs and miRNAs were identified between the cisplatin resistant and sensitive groups. A total of 139 DEGs in the resistant group were identified, where these included 23 downregulated and 116 upregulated genes (Figure 2B). Similarly, a total of 334 differentially expressed miRNAs (243 upregulated and 91 downregulated; Figure 2C) were identified in the resistant group.

Identification of differential AS events
To evaluate the correlation between AS events and treatment outcome, the level of AS events was analyzed. These data showed that that there were 1,711 AS events (665 upregulated and 1,046 downregulated) in the cisplatin resistant group (Figure 3A). In addition, we found a greater number and a higher percentage of alternate promoter (AP), but a lower number and a lower percentage of exon skip (ES) and retained intron (RI) in the upregulated AS events, compared with the downregulated AS events (Figure 3B,3C).
Construction of a prognostic signature for patients with LUSC and cisplatin treatment
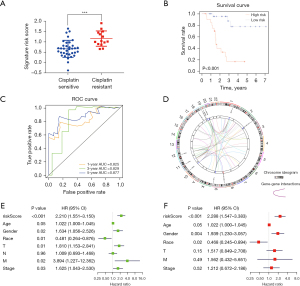
We next performed univariate Cox analyses of the aforementioned differential mRNAs, miRNA, and AS events to identify the candidate genes related to overall survival (OS). It was found that 14 mRNAs, 12 miRNAs, and 14 AS events significantly correlated with the OS of patients with LUSC with cisplatin treatment (Table S3). In addition, we carried out a LASSO Cox regression model analysis to identify key predictive biomarkers (Figure S1) and established an integrated prognostic model based on the expression of mRNA, miRNA, and the PSI value of AS events (Table 1). We calculated the risk score for each patient using the following formula: risk score = (0.33 × CTTN) + (−0.15 × CPM) + (−0.39 × hsa-miR-4746) + (0.29 × hsa-miR-4777) + (0.35 × hsa-miR-4664) + (−0.21 × has-miR-187) + (1.18 × VWA2|AT) + (−7.21 × ZNF675|AT) + (−4.22 × ZNF765|AT) + (11.10 × AIF1L|ES) + (2.30 × SELP|AT). In doing so, we found that the risk score was significantly higher (P<0.001) in the resistant group than in the sensitive group (Figure 4A). As shown in Figure 4B, Kaplan-Meier survival curves showed that patients with LUSC in the high-risk group had a significantly shorter OS (P<0.001) than those in the low-risk group, suggesting that this combined signature may be a useful tool for predicting the survival of patients. Next, we constructed ROC curves to assess the predictive efficiency of the model and found that the signature had a strong predictive ability, with high area under the curve (AUC) values of 0.825, 0.829, and 0.877 at 1, 3, and 5 years, respectively (Figure 4C). In addition, univariate and multivariate Cox regression analyses suggested that this signature was the best independent prognostic factor with the lowest P value (P<0.001) compared with the other potential prognostic factors (Figure 4D,4E). Furthermore, circus plots were constructed to illustrate the details of the AS events and their interacting genes in chromosomes (Figure 4F). The mutation status of these key genes was subsequently analyzed, where it was found that most of these genes had mutant sites, in particular genes with AS events (Figure S2).
Table 1
| Gene | Coefficient | P value |
|---|---|---|
| CTTN | 0.33 | <0.001 |
| CPM | −0.15 | 0.01 |
| hsa-miR-4746 | −0.39 | <0.001 |
| hsa-miR-4777 | 0.29 | 0.001 |
| hsa-miR-4664 | 0.35 | <0.001 |
| hsa-miR-187 | −0.21 | <0.001 |
| VWA2|13197|AT | 1.18 | 0.04 |
| ZNF675|48823|AT | −7.21 | 0.01 |
| ZNF765|51717|AT | −4.22 | 0.03 |
| AIF1L|87921|ES | 11.10 | 0.02 |
| SELP|8929|AT | 2.30 | 0.03 |
LUSC, lung squamous cell carcinoma.
Survival analysis of unstratified patients with LUSC using the integrated signature
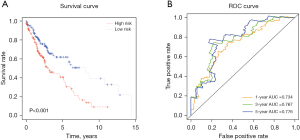
To determine whether the signature identified has predictive ability in unstratified patients with LUSC, Kaplan-Meier and ROC analyses were performed using the signature. As shown in Figure 5A,5B, the signature had strong predictive power (P<0.001) with high sensitivity and specificity for unstratified patients with LUSC. In addition, we also conducted stratified survival analyses to evaluate the predictive value of the integrated signature with various demographic and clinical parameters including age (<68 or ≥68 years), gender (male or female), stage (I + II or III + IV), T stage (T1 + T2 or T3 + T4), and N stage (N0 + N1 or N2). As shown in Figure 6, the signature showed significantly strong predictive ability for those who were male (P=0.001), aged <68 years (P<0.001), stage I + II (P<0.001), T1 + T2 (P<0.001), or N0 + N1 (P<0.001) disease; however, it showed no predictive value for other clinical parameters examined.
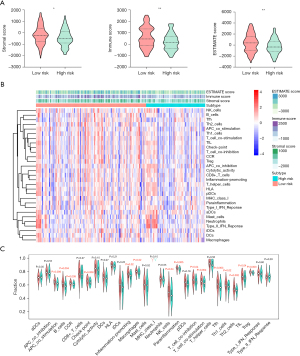
Correlation of signature with immune infiltration
To further explore the role of the prognostic signature in immunity, we analyzed the correlation of the signature with immune cell infiltration. The violin plots showed significantly higher stromal scores (P<0.05), immune scores (P<0.01), and ESTIMATE scores (P<0.01) in the low-risk group compared with the high-risk group (Figure 7A). In addition, using ssGSEA, the enrichment of 29 immune signatures was found in the two subgroups (Figure 7B). Furthermore, the results showed that patients in the low-risk group had higher levels of a range of immune cells than those in the high-risk group, including B cells and macrophages, amongst others (Figure 7C). These data suggest that the integrated signature can be used to identify immune characteristics and predict the immune microenvironment in LUSC.
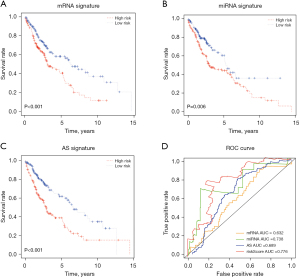
Superior prediction performance of the integrated signature
To demonstrate the superiority of predictive performance of our integrated signature, we compared the predictive values between our signature and the other published signatures (27-29). Although the Kaplan-Meier analysis showed that other mRNA, miRNA, and AS signatures also had significantly high prognostic value in patients with LUSC (Figure 8A-8C), the ROC analysis showed that our integrated signature had a higher AUC than the other mRNA, miRNA, and AS signatures from the literature (Figure 8D), suggesting that our integrated signature is indeed a better indicator for predicting the survival of patients with LUSC.
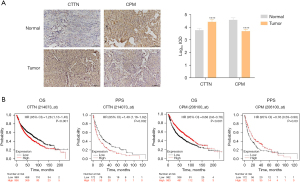
Diagnostic and prognostic potential of CTTN and CPM
To further validate the predictive value of this signature for LUSC, we used immunohistochemical staining to assess the protein levels of our two predictive biomarkers in lung tumour and normal lung tissues. Protein levels of CTTN were significantly higher in tumour samples (P<0.0001), whereas the protein levels of CPM were significantly lower in tumour lung tissue (P<0.0001) compared to normal lung tissue (Figure 9A). These data suggest that the signature has potential diagnostic utility for patients with LUSC. Additionally, using Kaplan-Meier plotter online tools, it was shown that patients with LUSC with higher CTTN levels had poor OS (P<0.001) and post progression survival (PPS) (P=0.002), whereas patients with higher CPM levels had a better OS (P<0.001) and PPS (P=0.03) (Figure 9B), suggesting that both biomarkers have prognostic value for patients with LUSC.
Small-molecule drug screening
There currently exists, an urgent unmet need to screen for novel drugs to combat cisplatin resistance in patients with LUSC. Therefore, we first performed differential analysis between the high-risk and low-risk groups and identified several differential genes. Subsequently, we inputted these genes into the CMap database to screen for potential small-molecule drugs. Ultimately, we identified three candidate small-molecule compounds targeting cisplatin resistance. These included sulforaphane, parthenolide, and BRD-K61033289. The 3D structures of each drug were downloaded from the PubChem database (Figure 10A). The IC50 concentration of sulforaphane and parthenolide was deduced in the SKMES1 LUSC cell line. As shown in Figure 10B, the IC50 for sulforaphane and parthenolide was 10.29±4.10 and 33.28±6.58 µM, respectively. In addition, using the GDSC database, the IC50 for parthenolide in two additional LUSC cell lines, NCI-H1869 and NCI-H226, was found to be 12.74 and 50.43 µM, respectively (Table 2).
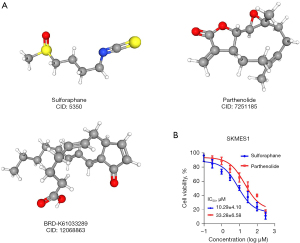
Table 2
| Cell lines | IC50 (μM) | Pathology |
|---|---|---|
| NCI-H1869 | 12.74 | Metastasis |
| NCI-H226 | 50.43 | Metastasis |
| SKMES1 | 33.28 | Metastasis |
IC50, the half-maximal inhibitory concentration; LUSC, lung squamous cell carcinoma.
Discussion
Chemotherapy a primary treatments modality for various cancers, including LUSC. Although treatment responses to cisplatin are initially effective, the clinical challenge of acquired drug resistance often results in treatment failure and relapse (30). The mechanisms of drug resistance are extremely complex and are typically associated with alterations in multiple pathways and genes. Therefore, there is an urgent need to identify biomarkers and novel molecular targets for cisplatin resistance in NSCLC, in particular patients with LUSC histology.
In recent years, numerous studies have reported that mRNA and miRNA may be involved in the drug resistance of various cancers, including lung cancer. For example, DEAD-box helicase 3 (DDX3) can mediate cisplatin resistance by regulating transcription factors in oral squamous cell carcinoma (31), RecQ like helicase 5 (RECQL5) can promote cisplatin resistance in NSCLC (32), miR-144 can target LIM homeobox 2 (LHX2) to reverse cisplatin resistance in cervical cancer (33), and miR-144-3p can mediate cisplatin resistance through targeting of NFE2 like BZIP transcription factor 2 (Nrf2) in lung cancer (30). Moreover, Liu et al. identified a 13-miRNA signature of cisplatin resistance, where these miRNAs could potentially target tumor protein P73 (TP73) and small ubiquitin like modifier 1 (SUMO1) in ovarian cancer cells (34). Fekete et al. identified specific miRNAs that are associated with resistance to platinum-based therapy in cervical, head and neck, and lung cancers (35). These studies point to the potentially critical role that mRNAs and miRNAs may play in reversing cisplatin resistance in cancer patients. In this study, we found that hsa-miR-187, which has been shown to alleviate cisplatin resistance in gastric cancer cells (36), was associated with a better prognosis in patients with cisplatin resistant LUSC. Although some of these mRNAs and miRNAs have not been reported as exerting a specific function in drug resistance, various reports have identified them as being involved in cell proliferation, apoptosis, and other biological processes in different cancer types. For instance, CTTN has been shown to promote cell proliferation and inhibit apoptosis in glioma (37), and hsa-miR-4746 has been associated with a better prognosis in LUSC (38), a finding that was verified in our study.
In recent years, a growing number of studies have demonstrated that AS events are promising biomarkers for the diagnosis and prognosis of various cancers including lung cancer. For example, increased ratio of insulin receptor isoform A (IR-A) and insulin receptor isoform B (IR-B) can reduce epithelial-mesenchymal transition (EMT) and prolong the survival of patients with LUSC (39). In addition, splicing factor SRSF1 was found to regulate BCL2 like 1 (Bcl-X) splicing and to interact with phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3) to suppress autophagy in lung cancer (40). Furthermore, accumulating evidence also supports the importance of AS events in chemoresistance. Krüppel-like factor 6 splice variant 1 (KLF6-SV1) for example, is an oncogenic splice variant of the KLF6 tumor-suppressor gene and can synergize with chemotherapeutic agents such as cisplatin resulting in tumor regression (41). Numerous genes are arranged in complex overlapping and interlaced patterns and such arrangements potentially contribute to the regulation of gene expression. In another study by Zhang et al., it was found that these overlapping genes including ERCC excision repair 1, endonuclease non-catalytic subunit (ERCC1), CD3E-associated protein (CD3EAP), and protein phosphatase 1 regulatory subunit 13 like (PPP1R13L) has potential applications for cisplatin resistance in NSCLC (42). MAD2gama, a novel mitotic arrest deficient 2 like 1 (MAD2) isoform, was found to be associated with cisplatin resistance in testicular germ cell tumors (43). Recent studies have shown that some AS events identified in our study, have a carcinogenic or suppressive role in different cancers. Aberrant expression of von Willebrand factor a domain containing 2 (VWA2) for example has been linked to the MYC proto-oncogene, BHLH transcription factor (MYC)-driven carcinogenic process in colorectal cancer (44). Moreover, Zhou et al. reported the downregulation of E2F transcription factor 1 (E2F1) which suppresses the migration and invasion of castration-resistant prostate cancer (CRPC) cells by regulating a number of key genes including tropomodulin 2 (TMOD2) and allograft inflammatory factor 1 like (AIF1L) (45). Selectin P (SELP) was also be shown to have considerable potential as a diagnostic biomarker in NSCLC (46). Moreover, Liu et al. reported a role for the insulin like growth factor 2 MRNA binding protein 2 (IGF2BP2)-F-box and leucine rich repeat protein 19 (FBXL19)-AS1-zinc finger protein 765 (ZNF765) feedback axis in the regulation of permeability of the blood-brain tumor barrier (BTB) and may serve as a potential therapeutic target in gliomas (47).
It is now well documented that the immune system is involved in controlling tumorigenesis and the progression of lung cancer (48), with immunotherapies now having an important therapeutic use in the treatment of NSCLC, in both LUAD and LUSC (49). In the current study, we found a higher stromal, immune and ESTIMATE score in the low-risk group. In addition, the distribution of several immune signatures was significantly different between the high-risk and the low-risk groups in LUSC, which included; APC co-stimulation, B-cells, C-C chemokine receptor (CCR), check-points, inflammation-promoting, macrophages, NK cells, parainflammation, T-cell co-inhibition, T-cell co-stimulation, Tfh, Th1 cells, Th2 cells, tumor-infiltrating lymphocytes (TILs) and regulatory T cells (Tregs). Abundant immune cell infiltration can increase the chance of tumor cells being attacked by the immune system, making the effect of immunotherapy more significant and the prognosis of patients better. These results demonstrated that immune escape or resistance to immunotherapy may, at least in part, account for drug resistance to cancer therapies. Importantly, we identified three small-molecule drugs (sulforaphane, parthenolide and BRD-K61033289) that have the potential to suppress cisplatin resistance in patients with LUSC. Among these three drugs, sulforaphane (4-methylsufinyl-3-butenyl isothiocyanate; SFE) is a traditional Chinese herbal medicine. A study support sulforaphane as a tumor suppressor in various cancers, including breast, ovarian, hepatocellular and lung cancers (50). Similarly, parthenolide could serve as a supplementary agent for suppressing various malignancies (51). In this study, we confirmed the inhibitory effects of sulforaphane and parthenolide in LUSC cells. However, the potential anti-cancer effects of BRD-K61033289 have not yet been reported. Therefore, the effects of these small-molecule drugs in cancer warrant further investigation.
In this study, we have constructed an integrated signature and identified several potential compounds, providing possibilities for future treatment of cisplatin-resistant LUSC patients. Furthermore, the predictive performance of this integrated signature is superior to that of individual mRNA, miRNA, and AS biomarkers. We acknowledge that this study is not without its limitations. The sample size was relatively small, consisting of 70 patients, and although the findings were verified in tissue samples, it is necessary to expand the sample size in further validation studies. In addition, mutated genes were not included in the multi-omics data as no differences between resistant and sensitive patients were found. Also, the clinical parameters examined did not include the smoking status or smoking history of patients, factors that will be taken into account in future studies considering the strong documented link between LUSC and smoking.
Conclusions
This study investigated the prognostic significance of mRNAs, miRNAs and AS events, from which we established an integrated signature for the prognosis of patients with LUSC with cisplatin resistance. These data add further knowledge to this important drug resistant phenotype in LUSC and may provide a valuable reference for exploring the underlying mechanisms and therapeutic targets of cisplatin resistance in LUSC.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-827/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-827/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-827/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-827/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Medical Ethics Committee of Liaoning Cancer Hospital (No. 20210826YG) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017;107:100-7. [Crossref] [PubMed]
- Derman BA, Mileham KF, Bonomi PD, et al. Treatment of advanced squamous cell carcinoma of the lung: a review. Transl Lung Cancer Res 2015;4:524-32. [Crossref] [PubMed]
- Chen WJ, Gan TQ, Qin H, et al. Implication of downregulation and prospective pathway signaling of microRNA-375 in lung squamous cell carcinoma. Pathol Res Pract 2017;213:364-72. [Crossref] [PubMed]
- Barr MP, Gray SG, Hoffmann AC, et al. Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS One 2013;8:e54193. [Crossref] [PubMed]
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 2014;740:364-78. [Crossref] [PubMed]
- Rudolph C, Melau C, Nielsen JE, et al. Involvement of the DNA mismatch repair system in cisplatin sensitivity of testicular germ cell tumours. Cell Oncol (Dordr) 2017;40:341-55. [Crossref] [PubMed]
- Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [Crossref] [PubMed]
- Guo L, Mohanty A, Singhal S, et al. Targeting ITGB4/SOX2-driven lung cancer stem cells using proteasome inhibitors. iScience 2023;26:107302. [Crossref] [PubMed]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010;463:457-63. [Crossref] [PubMed]
- Wahid M, Pratoomthai B, Egbuniwe IU, et al. Targeting alternative splicing as a new cancer immunotherapy-phosphorylation of serine arginine-rich splicing factor (SRSF1) by SR protein kinase 1 (SRPK1) regulates alternative splicing of PD1 to generate a soluble antagonistic isoform that prevents T cell exhaustion. Cancer Immunol Immunother 2023;72:4001-14. [Crossref] [PubMed]
- Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays 2014;36:236-43. [Crossref] [PubMed]
- Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov 2013;3:1228-37. [Crossref] [PubMed]
- Song J, Liu Y, Yin Y, et al. PTIR1 acts as an isoform of DDX58 and promotes tumor immune resistance through activation of UCHL5. Cell Rep 2023;42:113388. [Crossref] [PubMed]
- Pani T, Rajput K, Kar A, et al. Alternative splicing of ceramide synthase 2 alters levels of specific ceramides and modulates cancer cell proliferation and migration in Luminal B breast cancer subtype. Cell Death Dis 2021;12:171. [Crossref] [PubMed]
- Gordon MA, Babbs B, Cochrane DR, et al. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog 2019;58:196-205. [Crossref] [PubMed]
- Wang J, Liu T, Wang M, et al. SRSF1-dependent alternative splicing attenuates BIN1 expression in non-small cell lung cancer. J Cell Biochem 2020;121:946-53. [Crossref] [PubMed]
- Zhang F, Wang H, Yu J, et al. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer 2021;20:6. [Crossref] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Sharma A, Singh P, Jha R, et al. Exploring the role of miR-200 family in regulating CX3CR1 and CXCR1 in lung adenocarcinoma tumor microenvironment: implications for therapeutic intervention. Sci Rep 2023;13:16333. [Crossref] [PubMed]
- Singh P, Sharma A, Kumar B, et al. Integrative multiomics and weighted network approach reveals the prognostic role of RPS7 in lung squamous cell carcinoma pathogenesis. J Appl Genet 2023;64:737-48. [Crossref] [PubMed]
- Tabassum G, Singh P, Gurung R, et al. Investigating the role of Kinesin family in lung adenocarcinoma via integrated bioinformatics approach. Sci Rep 2023;13:9859. [Crossref] [PubMed]
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 2012;28:2385-7. [Crossref] [PubMed]
- Singh P, Solanki R, Tasneem A, et al. Screening of miRNAs as prognostic biomarkers and their associated hub targets across Hepatocellular carcinoma using survival-based bioinformatics approach. J Genet Eng Biotechnol 2024;22:100337. [Crossref] [PubMed]
- Gao M, Kong W, Huang Z, et al. Identification of Key Genes Related to Lung Squamous Cell Carcinoma Using Bioinformatics Analysis. Int J Mol Sci 2020;21:2994. [Crossref] [PubMed]
- Gan Z, Zou Q, Lin Y, et al. Construction and validation of a seven-microRNA signature as a prognostic tool for lung squamous cell carcinoma. Cancer Manag Res 2019;11:5701-9. [Crossref] [PubMed]
- Liu Y, Jia W, Li J, et al. Identification of Survival-Associated Alternative Splicing Signatures in Lung Squamous Cell Carcinoma. Front Oncol 2020;10:587343. [Crossref] [PubMed]
- Yin Y, Liu H, Xu J, et al. miR 144 3p regulates the resistance of lung cancer to cisplatin by targeting Nrf2. Oncol Rep 2018;40:3479-88. [Crossref] [PubMed]
- Shriwas O, Priyadarshini M, Samal SK, et al. DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated m(6)A-demethylation of FOXM1 and NANOG. Apoptosis 2020;25:233-46. [Crossref] [PubMed]
- Xia HW, Zhang ZQ, Yuan J, et al. Human RECQL5 promotes metastasis and resistance to cisplatin in non-small cell lung cancer. Life Sci 2021;265:118768. [Crossref] [PubMed]
- Shi F, Su J, Liu Z, et al. miR-144 reverses cisplatin resistance in cervical cancer via targeting LHX2. J Cell Biochem 2019;120:15018-26. [Crossref] [PubMed]
- Liu M, Zhang X, Hu CF, et al. MicroRNA-mRNA functional pairs for cisplatin resistance in ovarian cancer cells. Chin J Cancer 2014;33:285-94. [Crossref] [PubMed]
- Fekete JT, Welker Á, Győrffy B. miRNA Expression Signatures of Therapy Response in Squamous Cell Carcinomas. Cancers (Basel) 2020;13:63. [Crossref] [PubMed]
- Zhu QL, Li Z, Lv CM, et al. MiR-187 influences cisplatin-resistance of gastric cancer cells through regulating the TGF-β/Smad signaling pathway. Eur Rev Med Pharmacol Sci 2019;23:9907-14. [Crossref] [PubMed]
- Su HY, Lin ZY, Peng WC, et al. MiR-448 downregulates CTTN to inhibit cell proliferation and promote apoptosis in glioma. Eur Rev Med Pharmacol Sci 2018;22:3847-54. [Crossref] [PubMed]
- Chen B, Gao T, Yuan W, et al. Prognostic Value of Survival of MicroRNAs Signatures in Non-small Cell Lung Cancer. J Cancer 2019;10:5793-804. [Crossref] [PubMed]
- Jiang L, Zhu W, Streicher K, et al. Increased IR-A/IR-B ratio in non-small cell lung cancers associates with lower epithelial-mesenchymal transition signature and longer survival in squamous cell lung carcinoma. BMC Cancer 2014;14:131. [Crossref] [PubMed]
- Lv Y, Zhang W, Zhao J, et al. SRSF1 inhibits autophagy through regulating Bcl-x splicing and interacting with PIK3C3 in lung cancer. Signal Transduct Target Ther 2021;6:108. [Crossref] [PubMed]
- DiFeo A, Martignetti JA, Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat 2009;12:1-7. [Crossref] [PubMed]
- Zhang G, Xue P, Cui S, et al. Different splicing isoforms of ERCC1 affect the expression of its overlapping genes CD3EAP and PPP1R13L, and indicate a potential application in non-small cell lung cancer treatment. Int J Oncol 2018;52:2155-65. [Crossref] [PubMed]
- López-Saavedra A, Ramírez-Otero M, Díaz-Chávez J, et al. MAD2γ, a novel MAD2 isoform, reduces mitotic arrest and is associated with resistance in testicular germ cell tumors. Cell Cycle 2016;15:2066-76. [Crossref] [PubMed]
- González B, Fece de la Cruz F, Samuelsson JK, et al. Epigenetic and transcriptional dysregulation of VWA2 associated with a MYC-driven oncogenic program in colorectal cancer. Sci Rep 2018;8:11097. [Crossref] [PubMed]
- Zhou Q, Wang C, Zhu Y, et al. Key Genes And Pathways Controlled By E2F1 In Human Castration-Resistant Prostate Cancer Cells. Onco Targets Ther 2019;12:8961-76. [Crossref] [PubMed]
- Xing S, Zeng T, Xue N, et al. Development and Validation of Tumor-educated Blood Platelets Integrin Alpha 2b (ITGA2B) RNA for Diagnosis and Prognosis of Non-small-cell Lung Cancer through RNA-seq. Int J Biol Sci 2019;15:1977-92. [Crossref] [PubMed]
- Liu X, Wu P, Su R, et al. IGF2BP2 stabilized FBXL19-AS1 regulates the blood-tumour barrier permeability by negatively regulating ZNF765 by STAU1-mediated mRNA decay. RNA Biol 2020;17:1777-88. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Luo W, Wang Z, Tian P, et al. Safety and tolerability of PD-1/PD-L1 inhibitors in the treatment of non-small cell lung cancer: a meta-analysis of randomized controlled trials. J Cancer Res Clin Oncol 2018;144:1851-9. [Crossref] [PubMed]
- Wu G, Yan Y, Zhou Y, et al. Sulforaphane: Expected to Become a Novel Antitumor Compound. Oncol Res 2020;28:439-46. [Crossref] [PubMed]
- Sztiller-Sikorska M, Czyz M. Parthenolide as Cooperating Agent for Anti-Cancer Treatment of Various Malignancies. Pharmaceuticals (Basel) 2020;13:194. [Crossref] [PubMed]
(English Language Editor: J. Gray)



