
Preliminary exploration of poor prognostic factor IL-33 and its involvement in perioperative immunotherapy in stage II–III lung squamous cell carcinoma: a retrospective cohort study
Highlight box
Key findings
• Interleukin-33 (IL-33) was identified as an independent prognostic factor for patients with stage II and III lung squamous cell carcinoma (LUSC).
What is known, and what is new?
• This study verified the correlation between IL-33 and a poor prognosis in stage II–III LUSC at the gene and protein expression levels.
• This study explored the potential role of IL-33 blockade combined with immune checkpoint inhibitors (ICIs) in perioperative immunotherapy for stage II–III LUSC patients.
What is the implication, and what should change now?
• This study suggested that IL-33 blockade combined with ICIs in perioperative immunotherapy may improve the effect of anti-tumor immunotherapy. However, further confirmation is required through relevant animal experiments and clinical trials.
Introduction
Lung cancer remains the leading cause of cancer-related morbidity and mortality worldwide, as highlighted by the 2022 Report on the Burden of Malignant Tumors in China released by the National Cancer Center, which indicates that lung cancer has the highest incidence and mortality rates among all cancers (1). Globally, the 5-year overall survival (OS) rate for lung cancer ranges from 10% to 20%, with a specific rate of 19.7% reported in China. Studies indicated that early diagnosis and treatment are crucial for improving the survival outcomes of lung cancer patients (2,3). Lung squamous cell carcinoma (LUSC), a common type of non-small cell lung cancer (NSCLC), accounts for approximately 25% of all NSCLC cases. The prognosis of LUSC can be particularly poor due to the high likelihood of metastasis, as illustrated in recent case reports highlighting the gene profiling of metastatic sites post-surgery (4,5). Given the aggressive nature of LUSC and the lack of targetable genetic alterations, there is an urgent need to identify molecular biomarkers that can predict prognosis and guide treatment strategies.
In recent years, several prognostic biomarkers have been identified in NSCLC, such as epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangements (6,7). These biomarkers have been instrumental in advancing targeted therapies and improving patient outcomes. However, these biomarkers are primarily associated with lung adenocarcinoma (LUAD) rather than LUSC, highlighting a significant gap in the identification of reliable prognostic markers for LUSC (8). Previous study has also been limited by small sample sizes and a lack of comprehensive analysis across different NSCLC subtypes, resulting in inconclusive findings, especially in the context of LUSC (9). Furthermore, recent bioinformatics analyses have revealed distinct molecular profiles in LUSC compared to LUAD, further underscoring the need for specific research focused on LUSC biomarkers (10).
Interleukin-33 (IL-33) is a member of the IL-1 cytokine family and has been shown to play a complex role in tumor biology, influencing tumor transformation, growth, and metastasis in various malignant tumors, including NSCLC (11). IL-33 can exert its effects either by directly acting on tumor cells or indirectly affecting immune cells and tumor microenvironment. As is reported, the tumor microenvironment plays a critical role in the progression and prognosis of LUSC, with recent studies correlating various microenvironment-related markers to clinical outcomes (12). A study has shown that serum IL-33 levels are significantly higher in NSCLC patients than both healthy volunteers and individuals with benign lung diseases, and IL-33 serves as an independent prognostic factor for NSCLC (13). However, Yang et al. (14) found that lower IL-33 expression is associated with a poorer prognosis in LUAD. To date, there are no clear reports on relevant studies of IL-33 in LUSC, indicating a gap that this study aims to address.
Currently, multidisciplinary treatment combined with individualized treatment is recommended for the treatment of lung cancer (15). In recent years, clinical trials focusing on perioperative immunotherapy, or perioperative immunotherapy combined with chemotherapy for resectable stage II–III NSCLC have yielded promising preliminary results, offering hope for surgical intervention and the prognosis of locally advanced patients (16,17). This study retrospectively analyzed the prognostic role of IL-33 in stage II–III LUSC at both the gene transcription and protein expression levels, using clinical data from patients who underwent surgical resection. The study further explored the correlation between IL-33 and programmed death ligand 1 (PD-L1) expression, aiming to identify potential new targets for perioperative immunotherapy in stage II–III LUSC patients. We present this article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1122/rc).
Methods
Clinical data and experimental methods
Clinical data
This retrospective cohort study included data from 103 patients who underwent surgical resection of LUSC at Tianjin Medical University Cancer Institute & Hospital from November 1, 2004, to November 30, 2006. These patients had not received any anti-tumor treatments, such as chemotherapy, radiotherapy, targeted therapy, or immunotherapy before surgery. All patients were pathologically confirmed to have stage II–III LUSC postoperatively, with a total follow-up time exceeding 60 months. The primary endpoint of the study was OS.
The 103 patients were divided into two main groups for analysis. In the first group, the relationship between IL-33 expression and prognosis was evaluated in 83 patients. These patients were further randomly assigned to either a gene expression group (group A, 38 patients) or a protein expression group (group B, 45 patients). The remaining 20 patients (group C) were used for correlation analysis and Kaplan-Meier survival analysis of IL-33, PD-L1, and Ki-67 expression based on IHC results.
This research was approved by the Ethics Committee of Tianjin Medical University (No. Ek2024055) and was conducted in accordance with the principles outlined in the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients. Tissue samples were rapidly frozen and stored at −80 °C for subsequent analysis.
Sample size estimation and follow-up procedures
Sample size estimation was conducted based on prior studies suggesting that IL-33 might be a significant prognostic factor in NSCLC. We calculated that a minimum sample size of 85 patients would be necessary to detect a meaningful difference in survival outcomes with a power of 0.80 and an alpha level (α) of 0.05. Follow-up procedures included regular clinical evaluations, imaging studies, and laboratory tests at 3-month intervals during the first 2 years post-surgery, followed by 6-month intervals until 5 years post-surgery.
Assessment of baseline clinical factors and measurement of prognosis outcome
Baseline clinical factors such as sex, age, smoking history, and tumor stage were assessed at the time of diagnosis. Prognostic outcome was measured in terms of OS, defined as the time from the date of surgery to the date of death from any cause.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis for gene expression
Total RNA was extracted using the TRIzol kit (Thermo, Grand Island, NY, USA) according to the manufacturer’s instructions. The concentration and purity of the RNA was detected by UV-1800 ultraviolet spectrophotometer (Shimadzu, Kyoto, Japan), and the total RNA contents were calculated. A Molony murine leukemia virus (M-MLV) reverse transcription kit (Promega, Madison, WI, USA) was used to reverse transcribe the RNA into complementary DNA (cDNA) according to the instructions. An equal amount of RNA (1 µg) was reverse transcribed into cDNA for detection in the SYBR Premium Ex TaqTM 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The primers were synthesized by Santa Cruz (Dallas, TX, USA), and their sequences are listed in Table 1. The reverse transcription reaction mixture was 20 µL in total, comprising 10 µL of 2× SYBR Green premix, 0.4 µL each of 10 µmol/L forward and reverse primers, 2 µL of cDNA, with the remainder made up of double-distilled water. The reaction conditions were as follows: pre-denaturation at 95 °C for 2 minutes, followed by denaturation at 95 °C for 45 seconds, annealing at 60 °C for 20 seconds for 40 cycles, and final extension at 72 °C for 5 minutes to terminate the reaction. All the experiments were conducted in triplicate. ΔCT and ΔΔCT were calculated as follows: ΔCT = CTtarget gene − CTreference gene; ΔΔCT = ΔCTsample − ΔCTsample average. The relative expression quantity of the messenger RNA was calculated by 2−ΔΔCT.
Table 1
| Gene names | Primer sequence (5'-3') | Segment length (bp) |
|---|---|---|
| IL-33 | Forward primer 5'-GTGACGGTGTTGATGGTAAGAT-3' | 94 |
| Reverse primer 5'-AGCTCCACAGAGTGTTCCTTG-3' | ||
| β-actin | Forward primer 5'-GAGCACAGAGCCTCGCCTTT-3' | 129 |
| Reverse primer 5'-GGTGAGCTGCGAGAATAGCC-3' |
RT-qPCR, real-time quantitative polymerase chain reaction; IL-33, interleukin-33.
Immunohistochemistry (IHC) for protein expression
The streptavidin peroxidase method for IHC was used to detect the protein expression of IL-33 in the LUSC tissues. The paraffin-embedded tissue sections were sliced at a thickness of 4 µm. After baking at 65 °C for 20 minutes and deparaffinization in xylene, all the samples were rehydrated in graded alcohol and then blocked with 30 mL/L H2O2. Antigens were retrieved with heated citrate buffer (pH 6.0) for more than 20 minutes. The samples were blocked with rabbit anti-human serum. Subsequently, the sections were incubated overnight at 4 °C with appropriately diluted primary antibodies: IL-33 (1:200; bs-2633R; Bioss, Woburn, MA, USA); Ki-67 (1:200; bs-23103R; Bioss); PD-L1 (1:200; bs-22022R; Bioss). Following this, sections were incubated at room temperature for 30 minutes with secondary mouse anti-rabbit antibody labeled with streptavidin-horseradish peroxidase (HRP) (1:1,000). Finally, 3,3'-diaminobenzidine (DAB) staining kit (Polymer, Mentor, OH, USA) was used for visualization.
Under a light microscope, five representative high-power fields (400× magnification) were selected for each section and observed, and yellow to brownish-yellow or dark brown indicated a positive reaction.
Method for determining IL-33 IHC results
The positive rate referred to the percentage of positively stained cells in the cancer tissues, and was calculated as follows: 0–10%, a score of 0; 11–25%, a score of 1; 26–50%, a score of 2; 51–75%, a score of 3; and >75%, a score of 4.
In relation to the staining intensity score, the positively stained cells in the LUSC samples were categorized into the following four ranks based on their staining intensity: negative (which was scored as 0) for unstained cells; weakly positive (which was scored as 1) for cells showing a light yellow stain; moderately positive (which was scored as 2) for cells exhibiting a brownish-yellow stain; and strongly positive (which was scored as 3) for cells displaying a dark brown stain.
In relation to the results, the following formula was used: total score of each post-stain section = score of intensity of staining for that section × the number of staining positive cells for that section. A final score of ≤5 was defined as low expression and a final score >5 was defined as high expression (18).
Method for determining the Ki-67 IHC results
Five high-power fields were randomly selected for each slide, and the cell positive rate was calculated as follows: 0–5%: a score of 1; 6–50%: a score of 2; 51–75%: a score of 3; and 76–100%: a score of 4. A final score of ≤2 was defined as low expression, and a final score of >3 was defined as high expression (19).
Method for determining PD-L1 IHC results
Scoring was based on the proportion of positive cells. The positive cell proportion score involved randomly selecting five high-power fields (400×) under a light microscope, calculating the percentage of tumor cells showing positive staining in each field relative to all the tumor cells present, and using the average percentage across fields as the positive cell percentage. Based on the proportion of positive cells, the cells were scored as follows: no tumor cells stained: a score of 0; 1–49% stained, a score of 1; and >50% stained, a score of 2. A final score of 0–1 was defined as low expression, and a final score of 2 was defined as high expression (20).
Statistical analysis
Data were analyzed using SPSS 20.0 (IBM Corp., Armonk, NY, USA). Continuous variables were presented as median with interquartile ranges, and categorical variables were presented as frequencies and percentages. The hierarchical data were tested using a rank-sum test. The survival curve was drawn using the Kaplan-Meier method, and the survival analysis was performed using the log-rank test. Pearson correlation analysis was used to evaluate the relationships among the groups. The independent prognostic role of IL-33 (both gene and protein expression) in OS was assessed using multivariate logistic regression analysis. The regression model included IL-33 expression, tumor-node-metastasis (TNM) stage, T stage, and N stage as independent variables. Statistical significance was set at P<0.05, and all tests were two-sided.
Results
Relationship between IL-33 expression and clinicopathological characteristics in patients with stage II–III LUSC
IL-33 gene and protein expression in the LUSC tissues were analyzed to determine their correlation with the clinicopathological characteristics of the patients. The study included 103 patients, with 38 patients in the IL-33 gene expression group and 45 patients in the IL-33 protein expression group, all of whom had complete medical records. The analysis revealed no significant correlation between IL-33 expression (both gene and protein levels) and the gender, age, and smoking status of the patients (P>0.05 for all comparisons). However, a statistically significant correlation was observed between IL-33 expression and TNM stage, T stage, and N stage (P<0.05) (Table 2).
Table 2
| Clinicopathological characteristics | IL-33 gene expression | IL-33 protein expression | |||||
|---|---|---|---|---|---|---|---|
| Low expression (n=16) |
High expression (n=22) |
P | Low expression (n=17) |
High expression (n=28) |
P | ||
| Gender, n (%) | 0.18 | 0.72 | |||||
| Male | 15 (93.75) | 17 (77.27) | 13 (76.47) | 20 (71.43) | |||
| Female | 1 (6.25) | 5 (22.73) | 4 (23.53) | 8 (28.57) | |||
| Age (years), n (%) | 0.52 | 0.27 | |||||
| <60 | 12 (75.00) | 10 (45.45) | 7 (41.18) | 7 (25.00) | |||
| ≥60 | 4 (25.00) | 12 (54.55) | 10 (58.82) | 21 (75.00) | |||
| Smoking, n (%) | 0.17 | 0.26 | |||||
| No | 3 (18.75) | 1 (4.55) | 4 (23.53) | 3 (10.71) | |||
| Yes | 13 (81.25) | 21 (95.45) | 13 (76.47) | 25 (89.29) | |||
| TNM staging, n (%) | 0.03 | 0.02 | |||||
| IIA–IIIA | 15 (93.75) | 14 (63.64) | 16 (94.12) | 18 (64.29) | |||
| IIIB | 1 (6.25) | 8 (36.36) | 1 (5.88) | 10 (35.71) | |||
| T staging, n (%) | 0.02 | 0.04 | |||||
| T1–T3 | 16 (100.00) | 16 (72.73) | 16 (94.12) | 19 (67.86) | |||
| T4 | 0 (0.00) | 6 (27.27) | 1 (5.88) | 9 (32.14) | |||
| N staging, n (%) | 0.02 | 0.049 | |||||
| N0–N1 | 12 (75.00) | 8 (36.36) | 13 (76.47) | 13 (46.43) | |||
| N2 | 4 (25.00) | 14 (63.64) | 4 (23.53) | 15 (53.57) | |||
IL-33, interleukin-33; LUSC, lung squamous cell carcinoma; TNM, tumor-node-metastasis.
Survival analysis of IL-33 in patients with stage II–III LUSC
Survival analysis was conducted to evaluate the relationship between IL-33 gene expression and OS in patients with stage II–III LUSC. In the IL-33 gene expression group (n=38), the median survival time for patients with high IL-33 gene expression group (n=22) was 28 months, while the low IL-33 gene expression group (n=16) did not reach the median survival time, indicating a significantly higher survival rate in the low expression group (P=0.006) (Figure 1A).
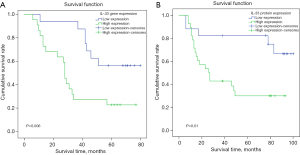
Similarly, in the IL-33 protein expression group (n=45), the high IL-33 protein expression group (n=28) had a median survival time of 26.3 months, whereas the low IL-33 protein expression group (n=17) did not reach the median survival time, demonstrating better survival outcomes in the low expression group (P=0.01) (Figure 1B).
Logistic regression analysis predicting prognostic factors in patients with LUSC
A multiple regression analysis was conducted to identify predictive variables using IL-33, TNM stage, T stage, and N stage as the independent variables, and OS as the dependent variable. The regression analysis revealed that both IL-33 gene expression and IL-33 protein expression were significant independent prognostic factors for OS. Specifically, the IL-33(gene expression) group had an F score of 7.761 (P<0.05), and the IL-33(protein expression) group had an F score of 4.414 (P<0.05). The overall significance test of the multiple linear regression indicated that the regression model was significant, which suggested that at least one independent variable would have an effect on the dependent variable. The stepwise regression analysis revealed that the independent risk factor affecting OS in stage II–III LUSC was IL-33 (P<0.05). Thus, in this study, the higher level of IL-33 in the cancer tissue of the stage II–III LUSC patients, the shorter the OS, which in turn indicated a poorer prognosis (Tables 3,4).
Table 3
| Prognostic factors | t | P |
|---|---|---|
| IL-33 gene expression | −2.138 | 0.04 |
| TNM | −1.802 | 0.08 |
| T | 0.260 | 0.80 |
| N | −1.907 | 0.07 |
LUSC, lung squamous cell carcinoma; IL-33, interleukin-33; TNM, tumor-node-metastasis.
Table 4
| Prognostic factors | t | P |
|---|---|---|
| IL-33 protein expression | −2.755 | 0.009 |
| TNM | −0.041 | 0.49 |
| T | −0.238 | 0.81 |
| N | −0.697 | 0.49 |
LUSC, lung squamous cell carcinoma; IL-33, interleukin-33; TNM, tumor-node-metastasis.
Survival analysis and correlation analysis for IHC of Ki-67, PD-L1, and IL-33
IHC staining results were shown in Figure 2. The relationship between protein expression of Ki-67, PD-L1, and IL-33 and survival time in IHC samples from 20 patients with stage II–III LUSC as demonstrated in Figure 3. The low expression groups for Ki-67, PD-L1, and IL-33 did not reach the median survival time. In contrast, the high expression groups had significantly shorter median survival times: 10 months for the Ki-67 high expression group (P=0.01), 9 months for the PD-L1 high expression group (P<0.001), and 16 months for the IL-33 high expression group (P=0.03).
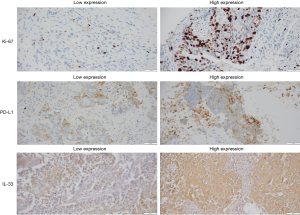
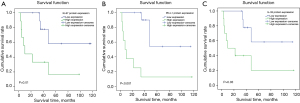
In the correlation analysis between IL-33 and the protein expressions of Ki-67 and PD-L1, the results demonstrated a positive correlation between IL-33 and Ki-67 protein expression (r=0.503, P=0.02), indicating statistical significance. Additionally, IL-33 and PD-L1 protein expression also exhibited a significant positive correlation (r=0.612, P=0.004). These results suggested that the expression levels of IL-33 may be correlated with those of Ki-67 and PD-L1 (Table 5).
Table 5
| Protein | IL-33 | |
|---|---|---|
| Pearson correlation coefficient | P value | |
| Ki-67 | 0.503 | 0.02 |
| PD-L1 | 0.612 | 0.004 |
IL-33, interleukin-33; PD-L1, programmed death ligand 1; LUSC, lung squamous cell carcinoma.
Discussion
In the context of LUSC, the identification of reliable biomarkers is crucial for improving diagnostic and prognostic accuracy. Recent studies, such as a research from Zhao et al. (21), have explored a range of potential biomarkers for LUSC, providing valuable insights into the molecular underpinnings of the disease. These efforts highlight the ongoing need to validate biomarkers that can be applied in clinical settings to guide treatment decisions. As is reported, IL-33 expression was identified as a poor prognostic factor in stage II and III LUSC (22). In this retrospective study, we used the RT-qPCR and IHC methods to validate its expression at both the gene transcription and protein translation levels, respectively. The high expression of both the IL-33 gene and protein significantly shortened the survival time of patients. Further, the high expression of IL-33 at both the gene and protein levels was significantly associated with advanced tumor T stage, N stage, and TNM stage, suggesting a poor prognosis. The multiple stepwise regression suggested that IL-33 was probably an independent risk factor for a poor prognosis of LUSC, which is consistent with relevant literature reports.
IHC research has shown that IL-33 is more highly expressed in LUAD and LUSC tissues than normal tissues, suggesting that IL-33 may be involved in tumor progression (23). As a cancer promoter, IL-33 increases tumor invasiveness in head and neck squamous cell carcinoma (24) and oral squamous cell carcinoma (25). The high expression of IL-33 is associated with poorer prognosis and shorter survival time, which suggests that IL-33 promotes the progression of squamous cell carcinoma. Additionally, studies on other malignant tumors, such as colorectal cancer, gastric cancer (26), and breast cancer (27), have also suggested that the high IL-33 expression promotes tumor invasion and metastasis, which is positively correlated with tumorigenesis and tumor progression. In our study, through IHC analysis of tumor tissues from LUSC patients, we also found that patients with high expression of IL-33 had significantly shorter survival periods compared to those with low expression.
Ki-67, also known as MKi67, is used to assess cell proliferation activity and differentiation potential. Previous studies have shown its association with the occurrence, progression, metastasis, and prognosis of various cancers (28,29). PD-L1, on the other hand, is involved in the immune evasion mechanism of NSCLC, particularly focusing on the alterations in PD-L1 expression from precancerous lesions to invasive cancer stages and its implications for immunotherapy. It has been observed that heightened PD-L1 expression facilitates tumor cell immune evasion (30). In our survival analysis, patients with high expression of Ki-67, and PD-L1 had significantly shorter survival periods compared to those with low expression. Furthermore, we observed a positive correlation between IL-33 expression and Ki-67 protein expression, as well as between IL-33 expression and PD-L1 expression.
Currently, surgery remains the primary treatment for early-stage NSCLC, but the indications for surgery in stage T1–3N2M0 (i.e., stage IIIA–IIIB) are still greatly disputed. However, the KEYNOTE-671 study revealed that pembrolizumab combined with chemotherapy significantly improved progression-free survival and pathological response outcomes in early resectable NSCLC patients compared to chemotherapy alone (31,32). In the meanwhile, feasibility and safety evaluations in neoadjuvant/perioperative immunotherapy clinical trials are ongoing. Wang et al. (33) have shown that IL-33 is a cancer-promoting factor in NSCLC cell growth and metastasis. Blocking IL-33 effectively inhibited the growth of NSCLC xeno-transplanted tumors in immunodeficient mice; thus, blocking IL-33 may be a promising method for treating NSCLC. Yue et al. (34) found that the simultaneous administration of IL-33 and programmed cell death protein 1 (PD-1) blockers significantly enhanced the efficacy of anti-PD-1 therapy in LUAD-bearing mice, showing better tumor control compared to anti-PD-1 alone. In the melanoma mouse model, PD-1 blockade combined with the cytokine IL-33 significantly inhibited tumor progression and improved their survival rates without immune-related adverse events, demonstrating the efficacy and safety of combination therapy, and lays the foundation for preclinical research (35). In mouse models of 4T1 breast cancer and CT26 colon cancer, the use of the co-blockade of the IL-33/ST2 and PD-L1/PD-1 axes was more effective in preventing tumor progression than a single blockade of either axis, which could provide a potential new approach to tumor immunotherapy (36). These findings highlight the potential of IL-33 in clinical treatment.
Although this study has produced significant findings, it is also important to acknowledge its various limitations. Firstly, the samples used in this study were relatively old, lacking modern positron emission tomography (PET) computed tomography (CT) scanning equipment in the early stages, which may have led to underestimation of disease staging in some patients. Furthermore, prolonged sample storage time may have caused protein degradation and changes in gene expression, potentially introducing bias into the experimental data. Secondly, the sample size was relatively limited, which may affect the generalizability and statistical significance of the findings, further validation of our results in larger cohorts is needed. Thirdly, we did not differentiate between survival outcomes among patients with stage II, IIIA, and IIIB, which may have limited a comprehensive understanding of the role of IL-33 across different disease stages. Finally, although we observed an association between IL-33 and the prognosis of LUCS patients, the specific biological mechanisms underlying this relationship require further investigation.
Conclusions
In summary, RT-qPCR and IHC techniques were used to validate molecular markers at the transcriptional and translational levels. The statistical analysis identified IL-33 as an independent prognostic factor for patients with stage II and III LUSC. It is highly likely that IL-33 will become a novel target for immunotherapy in LUSC. This study introduced the known prognostic adverse factors of Ki-67 and PD-L1 to further confirm the involvement of IL-33 in the malignant proliferation of tumor cells, and also suggested that IL-33 blockade combined with immune checkpoint inhibitors (ICIs) in perioperative immunotherapy could improve the effect of anti-tumor immunotherapy. However, further confirmation is required through relevant animal experiments and clinical trials.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1122/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1122/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1122/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1122/coif). N.S. obtained research grants from Eli Lilly, Chugai Pharmaceutical, Taiho Pharmaceutical, Pfizer, Ono Pharmaceutical, Nippon Kayaku, Takeda Pharmaceutical, Eisai, Shionogi, Daiichi Sankyo, and Boehringer Ingelheim and received speaking honoraria from Eli Lilly, AstraZeneca, MSD, Chugai Pharmaceutical, Taiho Pharmaceutical, Pfizer, Ono Pharmaceutical, Nippon Kayaku, Takeda Pharmaceutical, Daiichi Sankyo, Boehringer Ingelheim, Novartis, Kyowa Kirin, and Bristol Myers Squibb. W.M.B. received honoraria for lectures or educational events from AstraZeneca, Boehringer, Novartis, MSD, BMS, Lilly, Pfizer and Roche, received support for attending meetings and/or travel from Boehringer, Roche Pharma, and AstraZeneca, served on advisory board of Astra Zeneca, Boehringer, Novartis, MSD, Lilly Pharma, BMS, and Roche, and received equipment, material, drugs, medical writing, gifts or other services from Boehringer for medical writing. W.M.B.’s patent application EP21183549.1 (method for predicting a clinical response towards an immune checkpoint inhibitor based on pretreatment therewith) was filed with regard to the results of this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This work was performed according to the Helsinki Declaration (as revised in 2013) and was approved by the Institutional Ethics Committee of Tianjin Medical University (No. Ek2024055). Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zheng RS, Chen R, Han BF, et al. Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi 2024;46:221-31. [PubMed]
- He J, Li N, Chen WQ, et al. China guideline for the screening and early detection of lung cancer(2021, Beijing). Zhonghua Zhong Liu Za Zhi 2021;43:243-68. [PubMed]
- Dagher S, Sulaiman A, Bayle-Bleuez S, et al. Spread Through Air Spaces (STAS) Is an Independent Prognostic Factor in Resected Lung Squamous Cell Carcinoma. Cancers (Basel) 2022;14:2281. [Crossref] [PubMed]
- Miao JL, Liu RJ, Zhou JH, et al. Fibroblast Growth Factor Receptor 1 Gene Amplification in Nonsmall Cell Lung Cancer. Chin Med J (Engl) 2016;129:2868-72. [Crossref] [PubMed]
- Zhu F, Lin J, Liao H, et al. Gene profiling of metastatic small intestinal squamous cell carcinoma after lung squamous cell carcinoma surgery: a case report. Transl Cancer Res 2022;11:967-72. [Crossref] [PubMed]
- Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol 2008;21:S16-22. [Crossref] [PubMed]
- Restrepo JC, Dueñas D, Corredor Z, et al. Advances in Genomic Data and Biomarkers: Revolutionizing NSCLC Diagnosis and Treatment. Cancers (Basel) 2023;15:3474. [Crossref] [PubMed]
- Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. [Crossref] [PubMed]
- Thunnissen E, Kerr KM, Herth FJ, et al. The challenge of NSCLC diagnosis and predictive analysis on small samples. Practical approach of a working group. Lung Cancer 2012;76:1-18. [Crossref] [PubMed]
- Pan M, Huang P, Li L, et al. Comprehensive bioinformatics analysis on exportins in lung adenocarcinoma and lung squamous cell carcinoma. J Thorac Dis 2023;15:1872-91. [Crossref] [PubMed]
- Casciaro M, Cardia R, Di Salvo E, et al. Interleukin-33 Involvement in Nonsmall Cell Lung Carcinomas: An Update. Biomolecules 2019;9:203. [Crossref] [PubMed]
- Sugai M, Yanagawa N, Shikanai S, et al. Correlation of tumor microenvironment-related markers with clinical outcomes in patients with squamous cell carcinoma of the lung. Transl Lung Cancer Res 2022;11:975-90. [Crossref] [PubMed]
- Xie Q, Wang SC. IL-33, an important biomarker in non-small-cell lung cancer? Asian Pac J Cancer Prev 2013;14:7763. [Crossref] [PubMed]
- Yang M, Feng Y, Yue C, et al. Lower expression level of IL-33 is associated with poor prognosis of pulmonary adenocarcinoma. PLoS One 2018;13:e0193428. [Crossref] [PubMed]
- Travis WD, Dacic S, Wistuba I, et al. IASLC Multidisciplinary Recommendations for Pathologic Assessment of Lung Cancer Resection Specimens After Neoadjuvant Therapy. J Thorac Oncol 2020;15:709-40. [Crossref] [PubMed]
- Felip E, Altorki N, Zhou C, et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet 2021;398:1344-57. [Crossref] [PubMed]
- Ruck JM, Broderick SR. Neoadjuvant and Adjuvant Pembrolizumab for the Treatment of Early-Stage Resectable Non-small Cell Lung Cancer An Editorial Regarding the Interim Data Analysis of the KEYNOTE-671 Phase III Trial of Neoadjuvant and Adjuvant Pembrolizumab. Ann Surg Oncol 2024;31:4-5. [Crossref] [PubMed]
- Huang RSP, Haberberger J, Murugesan K, et al. Clinicopathologic and genomic characterization of PD-L1-positive uterine cervical carcinoma. Mod Pathol 2021;34:1425-33. [Crossref] [PubMed]
- van Zummeren M, Leeman A, Kremer WW, et al. Three-tiered score for Ki-67 and p16(ink4a) improves accuracy and reproducibility of grading CIN lesions. J Clin Pathol 2018;71:981-8. [Crossref] [PubMed]
- Xue J, Zhou A, Wu Y, et al. miR-182-5p Induced by STAT3 Activation Promotes Glioma Tumorigenesis. Cancer Res 2016;76:4293-304. [Crossref] [PubMed]
- Zhao X, Yuan C, He X, et al. Identification and in vitro validation of diagnostic and prognostic biomarkers for lung squamous cell carcinoma. J Thorac Dis 2022;14:1243-55. [Crossref] [PubMed]
- Choi MR, Sosman JA, Zhang B. The Janus Face of IL-33 Signaling in Tumor Development and Immune Escape. Cancers (Basel) 2021;13:3281. [Crossref] [PubMed]
- Xu J, Tang JL, Sun HY, et al. Study of IL-33 promoting EMT in non-small cell lung cancer. Journal of Clinical Pulmonary Medicine 2024;29:249-256.
- Peng L, Sun W, Chen L, et al. The Role of Interleukin-33 in Head and Neck Squamous Cell Carcinoma Is Determined by Its Cellular Sources in the Tumor Microenvironment. Front Oncol 2020;10:588454. [Crossref] [PubMed]
- Zhao M, He Y, Zhu N, et al. IL-33/ST2 signaling promotes constitutive and inductive PD-L1 expression and immune escape in oral squamous cell carcinoma. Br J Cancer 2023;128:833-43. [Crossref] [PubMed]
- Chang CP, Hu MH, Hsiao YP, et al. ST2 Signaling in the Tumor Microenvironment. Adv Exp Med Biol 2020;1240:83-93. [Crossref] [PubMed]
- Stojanovic B, Gajovic N, Jurisevic M, et al. Decoding the IL-33/ST2 Axis: Its Impact on the Immune Landscape of Breast Cancer. Int J Mol Sci 2023;24:14026. [Crossref] [PubMed]
- Klæstad E, Opdahl S, Raj SX, et al. Long term trends of breast cancer incidence according to proliferation status. BMC Cancer 2022;22:1340. [Crossref] [PubMed]
- Pergolini I, Crippa S, Pagnanelli M, et al. Prognostic impact of Ki-67 proliferative index in resectable pancreatic ductal adenocarcinoma. BJS Open 2019;3:646-55. [Crossref] [PubMed]
- Laville D, Casteillo F, Yvorel V, et al. Immune Escape Is an Early Event in Pre-Invasive Lesions of Lung Squamous Cell Carcinoma. Diagnostics (Basel) 2020;10:503. [Crossref] [PubMed]
- Provencio M, Nadal E, González-Larriba JL, et al. Perioperative Nivolumab and Chemotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med 2023;389:504-13. [Crossref] [PubMed]
- Wakelee H, Liberman M, Kato T, et al. Perioperative Pembrolizumab for Early-Stage Non-Small-Cell Lung Cancer. N Engl J Med 2023;389:491-503. [Crossref] [PubMed]
- Wang K, Shan S, Yang Z, et al. IL-33 blockade suppresses tumor growth of human lung cancer through direct and indirect pathways in a preclinical model. Oncotarget 2017;8:68571-82. [Crossref] [PubMed]
- Yue J, Guo H, Xu P, et al. Combination of IL-33 with PD-1 blockade augment mILC2s-mediated anti-tumor immunity. Cancer Immunol Immunother 2024;73:65. [Crossref] [PubMed]
- He H. Study on the Mechanism of PD-1 McAb Combined with Cytokine IL-33 in Antitumor Immune Response. Suzhou: Soochow University; 2018.
- Jovanovic MZ, Geller DA, Gajovic NM, et al. Dual blockage of PD-L/PD-1 and IL33/ST2 axes slows tumor growth and improves antitumor immunity by boosting NK cells. Life Sci 2022;289:120214. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


