
Predicting non-small cell lung cancer lymph node metastasis: integrating ctDNA mutation/methylation profiling with positron emission tomography-computed tomography (PET-CT) scan: protocol for a prospective clinical trial (LUNon-invasive Study)
Introduction
Lung cancer is the leading cause of cancer-related mortality, with non-small cell lung cancer (NSCLC) representing a significant portion, approximately 85%, of all lung cancer cases (1). Lymph node metastasis (LNM) in lung cancer manifests as an early occurrence in the metastatic process, preceding blood-borne metastasis (2-5). The presence of LNM strongly correlates with recurrence and survival rates, significantly influencing subsequent treatment strategies for lung cancer (2-5). Especially for patients diagnosed with operable NSCLC, the primary treatment option is often complete resection with systematic lymph node dissection (SLND). Nevertheless, determining whether SLND should be universally applied to all NSCLC patients undergoing surgery poses a significant dilemma. On the one hand, SLND may result in additional harm and complications (6). On the other hand, tumor-draining lymph nodes represent a critical source for the reinvigoration of antitumor immunity and a factor determining the treatment response to clinical immunotherapy (7-13). Therefore, preserving the unaffected tumor-draining lymph nodes is crucial (14,15). Overall, accurately predicting LNM in NSCLC before surgery is of utmost importance.
The mediastinal lymph node status is an important aspect of preoperative evaluation for operable lung cancer (stages I to III). This makes correct lymph node staging a crucial step before treatment for lung cancer patients. The most recommended methods include mediastinoscopy, endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA), and positron emission tomography-computed tomography (PET-CT). Among these, mediastinoscopy assessment is the gold standard, but it is highly invasive and costly, and its frequency of use in clinical practice is decreasing. EBUS-TBNA assessment is considered the next best option after mediastinoscopy. It is less invasive compared to mediastinoscopy and is becoming increasingly popular in clinical practice, gradually replacing mediastinoscopy. It is being recommended by various guidelines as the preferred method for lymph node staging in lung cancer (16). However, EBUS-TBNA is an invasive procedure with potential risks of complications and a certain proportion of false negatives [negative predictive value (NPV): 60–93%] (17-20). The use of PET-CT is the most common, with advantages including non-invasiveness and high safety. Generally, PET-CT is more accurate for lymph node staging compared to standalone computed tomography (CT). It has a sensitivity of 76%, a specificity of 88%, a negative likelihood ratio of 0.28, and a positive likelihood ratio of 6.4 (21). In a national registry study of patients with preoperative clinical stage cT1aN0M0 NSCLC, approximately 10% of patients were found to have mediastinal lymph node involvement confirmed by postoperative pathology (22). This is mainly because LNMs smaller than 4 mm may not be detected by PET-CT (22). Furthermore, up to 30% of pathologic stage T1N2 lung cancers have negative mediastinal lymph node status on preoperative PET-CT examination (23). Therefore, highly sensitive blood molecular markers combined with PET-CT are desired to improve the preoperative prediction of LNM (24,25).
LNM is the relay station for distant metastasis of lung cancer, which is directly proportional to the amount of circulating tumor cells in the blood, thereby showing a high potential to predict the occurrence of micrometastasis or occult LNM (26). Circulating tumor DNA (ctDNA) originates from apoptotic, necrotic, or secreted DNA fragments produced by tumor cells and is part of circulating cell-free DNA (cfDNA). The length of most cfDNA in healthy individuals ranges from 70 to 200 bp, but the length of ctDNA in cancer patients may be 200 bp or even exceed 1,000 bp. ctDNA contains the same genetic defects as the tumor DNA it originates from, such as point mutations, rearrangements, amplifications, microsatellite alterations, epigenetic modifications, etc. When tumor DNA enters the bloodstream, these genetic defect patterns can also be detected in plasma and serum. ctDNA has been widely used in the clinical diagnosis of driver mutations in advanced NSCLC (27). Minimal or molecular residual disease (MRD) refers to the detection of remaining tumor cells in the blood after initial therapy. Several groundbreaking studies have demonstrated the feasibility and potential applications of ctDNA in screening and detecting MRD after surgery for early-stage NSCLC (28-31). Recent data studies have shown that ctDNA levels are closely related to the staging of NSCLC, with higher levels in later-stage patients (32,33). Patients with operable lung cancer accompanied by mediastinal LNMs belong to the locally advanced stage, so it is expected that ctDNA may be more easily detected in the peripheral blood of these patients. Although this field has made many advances in predicting postoperative recurrence, guiding postoperative adjuvant therapy, and assessing treatment response, there is currently a lack of clinical evidence to determine the feasibility of detecting peripheral blood ctDNA to predict LNM in NSCLC lung cancer.
Compared to the detection of specific mutations in ctDNA, DNA methylation occurs in almost all stages of cancer, especially during the process of tumor metastasis and adaptation to new environments (34,35). In contrast to detecting tumor-specific mutations, abnormal methylation of specific gene regions in ctDNA exhibits highly consistent characteristics. Characteristic methylation “fingerprints” offer valuable insights for early cancer diagnosis and staging, efficacy assessment, recurrence monitoring, and prognosis evaluation. Out of the 28 million cytosine-phosphate-guanine (CpG) dinucleotide sites in the human genome, 60–80% of cytosine residues undergo methylation, with CpG island enrichment significantly enhancing the methylation signal-to-noise ratio. Moreover, methylation alterations tend to cluster, imparting higher specificity compared to base mutations. Thus, ctDNA methylation detection technology holds unique advantages in tumor detection sensitivity. However, there is a notable absence of investigations into methylation profiles specific to LNMs.
One of the critical considerations regarding ctDNA tests revolves around whether they are tumor-informed or tumor-naïve (36). Tumor-informed assays entail sequencing tumor tissue samples before blood ctDNA monitoring, allowing for the development of a personalized ctDNA panel based on mutation information obtained from the tumor tissue. In contrast, tumor-naïve assays use a fixed panel without specific mutation data from primary tumor tissue. The selection between these testing strategies is influenced by various factors, including evidence from ongoing randomized clinical trials, cost-effectiveness, and the accessibility of tumor tissue. The tumor-informed approach may pose challenges for patients encountering difficulties in obtaining tumor tissue, potentially leading to longer turnaround times that could impact decisions regarding adjuvant therapy. Conversely, tumor-naïve strategies typically offer shorter turnaround times and reduced costs compared to tumor-informed approaches. Nevertheless, the advantage of tumor-naïve methods in facilitating large-scale screening of suspicious genes comes with a trade-off. Specifically, there is a decrease in test specificity and an increase in false-positive results, emphasizing the importance of carefully considering the trade-offs when selecting a ctDNA testing strategy. Therefore, tumor-naïve approaches are often complemented with multi-omics methods, such as ctDNA methylation or fragment-omics, to enhance performance in terms of specificity, sensitivity, and sequencing depth. Further investigations are needed to determine whether combining tumor-naïve assays with ctDNA methylation profiles specific to LNMs yields significant performance improvements in clinical settings.
In summary, accurate preoperative prediction of LNMs holds paramount importance for the precise management of operable NSCLC patients in stages I–III. This study aims to establish a comprehensive model for predicting NSCLC LNMs, utilizing non-invasive, high-sensitivity, and high-specificity ctDNA mutation/methylation molecular features, in combination with PET-CT imaging characteristics. A series of studies will be conducted based on this model. The findings of this study furnish robust evidence for the precise diagnosis and treatment of operable NSCLC patients in stages I–IIIB.
Methods
Research design
This is a prospective study conducted in three stages. The timeline of this study is summarized in Figure 1. The volume of blood drawn will be 30 mL each time. ctDNA somatic mutation/methylation profiling, sponsored by Kanghui Biotech Co., Ltd. (Liaoning, China), covers 218 genes (Table S1).
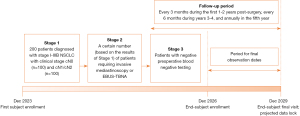
Stage 1
We will enroll 200 patients diagnosed with stage I–IIIB NSCLC who are scheduled for lobectomy and SLND. The prediction of LNM will be based on preoperative blood ctDNA mutation and methylation profiling. Specifically, during this stage, we will prospectively establish ctDNA methylation signatures specifically associated with LNMs in NSCLC. These molecular profiles will be combined with PET-CT imaging. Postoperative pathological results will serve as the reference standard for comparison.
Stage 1 entails the following details (Figure 2):
- Initial enrollment will include patients diagnosed with clinical stage I–IIIB NSCLC who have provided informed consent. Specifically, for stage I (c-N0) stage patients with ground glass nodules (≥2 cm) with a solid component, we focus on those requiring lobectomy, excluding cases that can be managed with sublobar resections).
- A highly sensitive tumor-naïve MRD panel of ctDNA will be employed to detect preoperative blood. Some patients will undergo methylation profiling to prospectively establish LNMs-specific ctDNA methylation signatures.
- Preoperative peripheral blood will be collected with separation of plasma and leukocytes and stored at −80 ℃ for future use.
- Immediately after surgery, surgical tissues should be collected, snap-frozen in liquid nitrogen, and stored at −80 ℃ for future use.
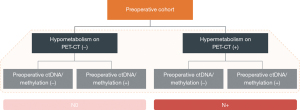
Building upon Stage 1, we will develop LNMs-specific ctDNA methylation signatures and assess the sensitivity and specificity of ctDNA mutation/methylation profiles alone or when combined with PET-CT scans for predicting LNMs. It is worth noting that particular attention will be given to the NPV. If we can achieve a significantly higher NPV using ctDNA alone or in conjunction with PET-CT, compared to clinical routine practices (such as mediastinoscopy, EBUS-TBNA, or PET-CT alone or in combination), this will ensure that the majority of cases with negative ctDNA results are indeed negative. We will then proceed to the third stage.
The data obtained from Stage 1 will inform the implementation of Step 2.
Stage 2
A certain number (sample size will be determined based on the results of Stage 1) of patients requiring invasive mediastinoscopy or EBUS-TBNA, based on the clinical guidelines, will be included for blood ctDNA mutation/methylation profile (or combined PET-CT), and the detection rate of LNMs will be assessed in patients with negative preoperative blood ctDNA mutation/methylation (or combined PET-CT). The accuracy of predicting LNMs will be compared with that of patients with negative or positive preoperative blood ctDNA mutation/methylation (or combined PET-CT).
Stage 2 entails the following details (Figure 3):
- Patients diagnosed with clinical stage I–IIIB NSCLC and necessitating invasive mediastinoscopy or EBUS-TBNA, as per clinical guidelines, will be enrolled upon providing informed consent.
- Preoperative peripheral blood will be collected, with plasma and leukocytes separated and stored at −80 ℃ for blood ctDNA mutation/methylation testing.
- The detection rate of LNMs will be evaluated in patients with negative preoperative blood ctDNA mutation/methylation (alone or combined with PET-CT).
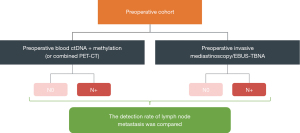
Building upon Stage 2, we will evaluate the efficacy of negative ctDNA mutation/methylation profiles alone or in conjunction with PET-CT scans for ruling out LNMs in patients undergoing mediastinoscopy or EBUS-TBNA. The objective is to determine if negative testing ctDNA somatic mutation/methylation profiles alone or in combination with PET-CT can achieve comparable or superior predictive performance compared to mediastinoscopy or EBUS-TBNA, potentially supplanting these conventional invasive methods.
The data obtained from Stages 1 and 2 will inform the implementation of Step 3.
Stage 3
For patients with negative preoperative blood negative testing (ctDNA mutation/methylation profiles alone or in combination with PET-CT), systematic lymph node sampling/dissection and no lymph node dissection will be grouped and compared. Regular follow-up will be performed to investigate the impact on long-term MRD negative duration and prognosis. The primary outcomes include postoperative MRD negative duration and disease-free survival (DFS); the second outcome would be the overall survival (OS).
Stage 3 entails the following details (Figure 4):
- Patients with operable NSCLC scheduled for lobectomy and who would provide an informed consent form for initial enrollment.
- Preoperative blood for ctDNA mutation/methylation profiles and PET-CT data will be collected.
- Patients with negative ctDNA mutation/specific methylation and PET-CT results may be included in this phase of the trial.
- Upon signing an informed consent form, patients will be categorized into two groups undergoing lymph node dissection and those not undergoing dissection. They will be followed up every 3 months for the first 2 years post-surgery, every 6 months for years 3–4, and annually for the fifth year.
- Primary outcomes will include postoperative MRD-negative duration and DFS.
- Secondary outcomes will encompass OS.
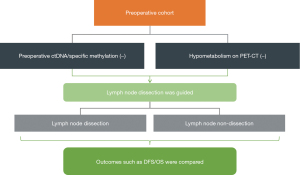
Stage 3 aims to assess whether a non-invasive approach, which integrates preoperative blood ctDNA mutation/methylation profiles and PET-CT data, can inform the intraoperative surgical resection of lymph nodes.
Follow-up
The follow-up period pertains to the third stage of the trial, during which patients will undergo follow-up according to designated postoperative intervals outlined in the study design. Specifically, patients will receive follow-up evaluations every 3 months during the first 1–2 years post-surgery, every 6 months during years 3–4, and annually in the fifth year. Follow-up records will encompass various aspects, including the general physical status of patients, disease recurrence or progression, their survival status, reasons for study withdrawal, updates on tumor site screening via enhanced chest CT, and documentation of disease progression timelines. Additionally, annual enhanced brain magnetic resonance imaging (MRI) and bone scans will be conducted.
Patients who discontinue participation or withdraw from the study for reasons unrelated to disease progression will still undergo tumor assessments every 6–12 months to monitor for objective disease progression. Following disease progression, patients will be contacted every 6–12 months by either their family members, primary physician, or the research team to ascertain their survival status.
Evaluation of LNMs
Assessment of LNMs primarily relies on anatomical lung lobectomy, which encompasses procedures such as single lobectomy, sleeve lobectomy, double lobectomy, and whole lung resection, along with SLND or sampling. Lymph node dissection involves stations 2–14.
CFI, DFS, OS
CFI is defined as the duration between a patient’s transition from ctDNA negative to ctDNA positive status.
DFS is defined as the duration from a patient’s surgical resection to tumor recurrence.
OS is defined as the time from patient surgical resection to death from any recorded cause.
Research purpose
Building upon Stage 1, we will develop LNMs-specific ctDNA methylation signatures and assess the sensitivity and specificity of ctDNA mutation/methylation profiles alone or when combined with PET-CT scans for predicting LNMs. It is worth noting that particular attention will be given to the NPV.
Building upon Stage 2, we will evaluate the efficacy of negative ctDNA mutation/methylation profiles alone or in conjunction with PET-CT scans for ruling out LNMs. The objective is to determine if negative testing (ctDNA mutation/methylation profiles alone or in combination with PET-CT) can achieve comparable or superior predictive performance compared to mediastinoscopy or EBUS-TBNA, potentially supplanting these conventional invasive methods.
Building upon Stage 3, we will aim to assess whether a non-invasive approach, which integrates preoperative blood ctDNA mutation/methylation profiles and PET-CT data, can inform the intraoperative surgical resection of lymph nodes.
Target study population
Patients diagnosed with surgically resectable NSCLC, clinically staged I–IIIB (where primary treatment is not required), who consent to surgery and are scheduled for anatomical lobectomy (including single lobe, sleeve resection, double lobe, or whole lung) with SLND or sampling, are eligible for inclusion.
Among these patients, those with clinical stage I ground-glass nodules (defined as ground-glass opacity >2 cm with a solid component in the lung, regardless of the proportion of solid component) will undergo anatomical lobectomy with SLND or sampling. The distribution of clinical stages in the enrolled population is as follows: clinical stage cN0 (n=100) and cN1/cN2 (n=100).
Selection criteria
Each subject should fulfill all the inclusion criteria for this study and not have any exclusion criteria.
Inclusion criteria
Patients meeting the following criteria may be eligible for inclusion as study participants:
- Provision of informed consent prior to study initiation.
- Age between 18 and 75 years.
- Preoperative clinical evaluation indicating NSCLC stage I–IIIB (without initial treatment sought).
- Newly treated patients who have consented to undergo surgery.
- Eastern Cooperative Oncology Group (ECOG) performance status (PS) score of 0–1.
Exclusion criteria
Participants will be excluded from the study if any of the following criteria are met:
- NSCLC histopathologically determined to be other than stage I–IIIB following surgical evaluation.
- Presence of other active malignant tumor diseases.
- Evidence of any serious or uncontrolled systemic illness, including uncontrolled hypertension and active bleeding, as assessed by the investigator, may contribute to reluctance to participate in the trial or decrease adherence to the study regimen. Additionally, active infectious diseases such as hepatitis B, hepatitis C, and human immunodeficiency virus (HIV) infection are grounds for exclusion.
- History of interstitial lung disease (ILD), drug-induced ILD, radiation pneumonitis requiring steroid therapy, or any indication of clinically active ILD.
- Patients considered by the investigator to be unable to comply with the study protocol, restrictions, and requirements, or those facing circumstances at the investigator’s discretion that would impede their participation in the study.
Procedures for patient withdrawal from the study
Patients who do not meet the eligibility criteria should under no circumstances be enrolled in this clinical study. Additionally, enrolled patients who are later discovered not to meet all eligibility criteria must be withdrawn from the study.
Termination and withdrawal from studies
Patients who voluntarily withdraw from the study, if their data is deemed valuable for evaluation, will not be replaced. However, if withdrawn patients do not provide valuable data, they may be replaced by new patients to maintain the minimum required number of assessable patients.
Patients may withdraw from the study at any time without affecting further treatment (withdrawal of consent). Investigators should ask these patients about their reasons for withdrawing. Researchers should inquire about the reasons for patient withdrawal. Whenever possible, researchers should conduct planned post-study assessments on patients before they are considered fully withdrawn from the study.
Research plan
The data collection timeline is summarized in Table 1.
Table 1
| Assessment/procedures | Inclusion screening (start of the study) | Stage 2 | ctDNA-negative patients follow-up period (every 3 months for the first 2 years; every 6 months for 3–4 years; every 12 months in the fifth year) | Disease progress |
|---|---|---|---|---|
| Informed consent | × | |||
| Baseline population characteristics | × | |||
| Clinical examination | × | |||
| ECOG status | × | |||
| ECG | × | |||
| Treatment history | × | |||
| Preoperative blood ctDNA | × | |||
| Preoperative blood methylation | × | |||
| Invasive mediastinoscopy/EBUS-TBNA | × | |||
| Surgical tissue | × | |||
| Inclusion exclusion review | × | |||
| Chest enhanced CT | × | × | ||
| Withdrawal and reasons | × | |||
| State of survival | × | × |
“×” indicates the items included in this stage of the study. Enrollment screening was applied to both Stage 1 and Stage 2 studies. Follow-up and disease progression were adapted only for the Stage 3 trial. ctDNA, circulating tumor DNA; ECOG, eastern cooperative oncology group; ECG, electrocardiogram; EBUS-TBNA, endobronchial ultrasound-guided transbronchial needle aspiration; CT, computed tomography.
Enrollment/screening period
Patients clinically diagnosed with stage I–IIIB NSCLC will be scheduled for anatomical lobectomy (including single lobe, sleeve resection, double lobe, and whole lung) with SLND or sampling. Among them, patients with ground-glass nodules in clinical stage I will be required to have ground-glass in the lung measuring more than 2 cm with solid components (regardless of the proportion of solid components). Enrollment will be open to patients with either positive or negative ctDNA results in preoperative blood tests. This period constitutes the enrollment/screening phase during which investigators will obtain signed informed consent from potential patients or their legally authorized representatives.
Key components of the enrollment/screening period include:
- Clinical staging distribution: the enrolled population will include patients with clinical staging cN0 (n=100) and cN1/cN2 stage (n=100).
- Baseline population characteristics: general patient characteristics, including age, gender, and race (if available or permitted by local regulations), will be collected.
- Physical examination: baseline physical examination findings will be recorded.
- ECOG PS: patients with an ECOG PS score of 0–1 will be eligible for enrollment.
- Electrocardiogram (ECG) recordings: ECG recordings will be obtained as indicated at baseline.
- Treatment history: patients must be treatment-naïve for lung cancer, with no history of related comorbidities or treatments, including chronic diseases requiring drug therapy.
- Tumor assessment: tumor histology, stage at diagnosis, and stage at study inclusion will be evaluated before and after tumor tissue resection.
- Laboratory examination: laboratory parameters such as blood routine, biochemistry, and liver and kidney function will be assessed. Any abnormalities will be documented in the case report form (CRF).
- ctDNA screening: preoperative blood samples, surgical tissue, and postoperative monitoring blood (collected 3 days after surgery) will be obtained for ctDNA screening.
- Serious adverse events (SAEs): monitoring for SAEs will commence upon patient enrollment and informed consent.
Research assessment
The CRF system will be used for data collection and queries. Investigators should ensure that data recorded in the CRF conform to the protocol provisions and instructions provided.
Researchers are responsible for ensuring the accuracy, completeness, and timeliness of data recording and must be able to provide data query results based on the clinical research protocol.
Definition of study endpoints
To achieve the objectives of this study, relevant data on the following study endpoints will be collected.
Primary endpoints
- Stage 1: Establishing LNMs-specific ctDNA methylation signatures; NPV rates of LNMs between different groups using preoperative blood ctDNA somatic mutation/methylation alone, or in combination with PET-CT, and PET-CT.
- Stage 2: Detection rates between ctDNA somatic mutation/methylation profiles alone or in combination with PET-CT and that of traditional mediastinoscopy/EBUS-TBNA.
- Stage 3: CFI and DFS between the presence and absence of systematic lymph nodes in patients with preoperative negative ctDNA profiling and/or PET-CT.
Second endpoints
- Stage 3: OS between the presence and absence of systematic lymph nodes in patients with preoperative negative ctDNA profiling and/or PET-CT.
Statistical analysis
All statistical analyses will be conducted using R or SAS 9.2 statistical software and following a predefined statistical analysis plan. Missing data is an inevitable problem in clinical trials. Due to the dropout of cases, the primary purpose (preoperative/postoperative ctDNA) or exploratory purpose (DFS/OS, etc.) of the study should be removed from the corresponding analysis, and only the complete data should be analyzed.
For comparison between the two groups, all hypothesis tests will be performed by a two-sided test (α =0.05). P<0.05 was considered statistically significant. All confidence intervals (CIs) will be given 95% confidence.
The demographic characteristics, general information and baseline data of the two groups should be described by simple statistical description and difference analysis. Continuous variables will be described by mean, standard deviation, median, maximum, and minimum. Two independent sample t-tests, the Welch or Behren-Fisher test, or the Wilcoxon rank-sum test will be used for the difference test. Categorical data will be used to describe the number and percentage of cases, and χ2 test or Fisher’s exact test will be used for the difference test.
Research data management
Sample size calculation
Based on previous studies, the sensitivity of ctDNA methylation detection is above 70% in stage I lung adenocarcinoma (37,38). In a study employing a machine learning model to detect circulating free DNA cfDNA from tumors, the integration of fragmentation features, clinical risk factors, and carcinoembryonic antigen (CEA) levels, supplemented by CT imaging, enabled the detection of 94% of cancer patients across various stages and subtypes. This included a detection rate of 91% in stage I/II and 96% in stage III/IV, with a specificity of 80% (39). Consequently, we assume that the sensitivity of the ctDNA assay ranged between 70% and 96% in our study. Since our study focuses on early-stage patients, we hypothesize a postoperative positive rate of 15% during the 5-year follow-up, with an 85% follow-up rate. Given this information, we plan to recruit 200 individuals for the final analysis. Consequently, with an alpha of 0.05, the two groups (postoperative positive vs. postoperative negative) are expected to demonstrate a statistically significant difference in DFS, with a power of over 90%.
Monitoring of research
During the study period, the investigative team from the testing party will maintain regular contact with the research centers, including accessing the following information:
- Providing information and support to the researchers.
- Ensuring facility operations remain normal.
- Ensure that the research team strictly adheres to the research protocol, data are accurately and timely recorded in the CRFs, and biological samples are processed according to the laboratory manual.
- Performing source data verification (comparing data in CRFs with subjects’ medical records at hospitals or practice centers, including other study-related records), including verification of subjects’ informed consent forms. This requires direct access to all original records (such as clinical charts) for each subject.
- Ensuring reporting of subjects’ withdrawal of consent for the use of biological samples, ensuring subjects are informed about the identification and disposition/destruction of their biological samples, and recording relevant operations.
If researchers or other research center personnel require information or advice related to study execution, they must be able to contact the investigative team from the testing party between visits.
Source data
The research center will maintain source data following the Good Clinical Practice (GCP; 2020) for drug trials regulations.
Research timeline and closure of the study
The following requirements should be met prior to patient participation in the study and the start of any study-related procedures. The testing party and the investigator/research center have signed a clinical study protocol and other clinical study agreements.
The program study timeline is projected below:
- Estimated first subject enrollment: December 2023.
- Estimated end-subject enrollment: December 2026.
- Estimated end-subject final visit: December 2029.
- Projected data lock: December 2029.
If the investigator decides to terminate or temporarily interrupt the study, a written document detailing the reasons for the termination or temporary interruption must be provided to the principal investigator and site management. The principal investigator must inform the patient immediately of this decision and provide appropriate treatment and appropriate measures to ensure patient safety and well-being.
Data processing
CRF should be used for this study. For each patient, a complete CRF signed by the principal investigator or his authorized investigator should be available. This principle also applies to recording the CRFs of patients who did not complete the trial (i.e., CRFs were filled out during screening). Investigators should ensure the accuracy, completeness, legality, and timeliness of all data and required reporting in the CRF and timely reporting to the testing parties.
Quality control
The aim is to control the quality of the testing procedures, monitor the analytical performance of the testing methods, alert personnel to existing problems, and ensure the accuracy and comparability of test results.
Moral and ethical regulatory requirements
Ethical statement
The study will be conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol and informed consent documents have been approved by the Ethical Committee of Shanghai Chest Hospital (approval No. IS23098). Informed consent will be obtained from all the patients.
In addition, the study will comply with the study protocol, the International Conference on Harmonisation Guidelines for Good Clinical Practice, and applicable local regulatory requirements and laws.
Subject data protection
Informed consent forms will include language compliant with relevant data protection and privacy legislation (or, in certain cases, may be accompanied by a separate document containing such content). The testing party shall not disclose personal genetic results to participants, any insurance companies, any employers, their family members, general practitioners, or any other third party unless required by law. Precautions will be taken to maintain confidentiality and prevent the linkage of genetic data with participant identities. However, in special circumstances, certain individuals may have access to both a participant’s genetic data and personal identity.
Discussion
In the TRACERx study (40), researchers utilized anchored multiplex polymerase (AMP) chain reaction enrichment panels to scrutinize the subclonal dynamics of lung cancer recurrence and metastasis. In 2017, this study reported a mean lead time of 70 days for ctDNA detection compared to radiological assessments. An update presented at the 2020 American Association for Cancer Research (AACR) revealed a lead time averaging 151 days (ranging from 0 to 984 days) from ctDNA detection to clinical relapse. In 2019, a study showed that ctDNA levels decreased rapidly after radical tumor resection, and ctDNA levels between 3 days and 1 month after surgery can be used as a baseline value for postoperative lung cancer monitoring (41). The recurrence-free survival of patients with ctDNA positive and negative 3 days after surgery was 278 and 637 days (P value: 0.002), respectively. Beyond monitoring relapse, ctDNA MRD testing holds promise in identifying high-risk patient cohorts likely to benefit from adjuvant therapy (42,43).
Considering the heterogeneity of patients with the same tumor, node, metastasis (TNM) stage (a system for classifying a malignancy which is based on assessing the tumor, regional lymph nodes, and distant metastasis), some individuals may undergo unnecessary adjuvant therapy, while others at high risk may defer essential treatments. Therefore, the detection of MRD via ctDNA testing may serve as a valuable adjunct to the TNM staging system (44). The TracerX study found a significantly higher preoperative blood ctDNA positivity rate in patients with N2 LNM compared to those with no lymph node involvement or proximal metastasis (N0/N1 group), at 79% vs. 29%, respectively. However, there is currently no research predicting LNM status in operable patients based on ctDNA. Recent studies have highlighted the close relationship between specific gene methylation and the development of malignant tumors, serving as novel molecular markers for tumor detection (45,46). Methylation detection in lung cancer serves as a highly specific and sensitive indicator for pulmonary disease diagnosis, enhancing the diagnosis of early-stage lung cancer. Previous research has reported that gene aberrant methylation leads to the downregulation of SOCS3 expression in NSCLC tissue and is closely associated with NSCLC LNM. If non-invasive ctDNA monitoring could provide a basis for patients to avoid unnecessary lymph node dissection, it would hold significant clinical value for the treatment modalities and long-term management of lung cancer.
Training of research center personnel
The laboratory research team will review and discuss the trial protocol requirements and related documentation with the trial investigators before the initiation of the study. The principal investigator is required to ensure that all investigators are appropriately trained and that participants are informed in advance of any updates regarding the conduct of the study. The principal investigator must keep records of all persons (doctors, nurses, and other staff) who participate in the study.
Strength of the study
The study is divided into three stages, with the results of each phase serving as the basis for the subsequent phase. If the results of this study are ideal, we can use the non-invasive, highly sensitive, and highly specific ctDNA mutation/methylation molecular signatures, combined with PET-CT imaging features, to develop an accurate integrated model for predicting LNMs in NSCLC.
Limitations of the study
This study is single-center research conducted without participation from other hospitals, potentially introducing bias in patient demographics and characteristics. Additionally, patients in the third stage will be followed up for a long time, and there may be problems such as data loss.
Acknowledgments
Funding: The ctDNA test in this study was supported by the Liaoning Kanghui Biotechnology Co., Ltd.
Footnote
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1033/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1033/coif). F.Y. reports that the ctDNA test in this study was supported by the Liaoning Kanghui Biotechnology Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study will be conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol and informed consent documents have been approved by the Ethical Committee of Shanghai Chest Hospital (approval No. IS23098). Informed consent will be obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Preoperative chemotherapy for non-small-cell lung cancer: a systematic review and meta-analysis of individual participant data. Lancet 2014;383:1561-71. [Crossref] [PubMed]
- Quinn JJ, Jones MG, Okimoto RA, et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 2021;371:eabc1944.
- Brown M, Assen FP, Leithner A, et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 2018;359:1408-11. [Crossref] [PubMed]
- Pereira ER, Kedrin D, Seano G, et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 2018;359:1403-7. [Crossref] [PubMed]
- Ma Q, Dieterich LC, Ikenberg K, et al. Unexpected contribution of lymphatic vessels to promotion of distant metastatic tumor spread. Sci Adv 2018;4:eaat4758. [Crossref] [PubMed]
- Cho HJ, Kim DK, Lee GD, et al. Chylothorax complicating pulmonary resection for lung cancer: effective management and pleurodesis. Ann Thorac Surg 2014;97:408-13. [Crossref] [PubMed]
- O'Melia MJ, Manspeaker MP, Thomas SN. Tumor-draining lymph nodes are survival niches that support T cell priming against lymphatic transported tumor antigen and effects of immune checkpoint blockade in TNBC. Cancer Immunol Immunother 2021;70:2179-95. [Crossref] [PubMed]
- Wu TD, Madireddi S, de Almeida PE, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020;579:274-8. [Crossref] [PubMed]
- Spitzer MH, Carmi Y, Reticker-Flynn NE, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 2017;168:487-502.e15. [Crossref] [PubMed]
- Prokhnevska N, Cardenas MA, Valanparambil RM, et al. CD8(+) T cell activation in cancer comprises an initial activation phase in lymph nodes followed by effector differentiation within the tumor. Immunity 2023;56:107-124.e5. [Crossref] [PubMed]
- Huang Q, Wu X, Wang Z, et al. The primordial differentiation of tumor-specific memory CD8(+) T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell 2022;185:4049-4066.e25. [Crossref] [PubMed]
- Yang H, Sun B, Ma W, et al. Multi-scale characterization of tumor-draining lymph nodes in resectable lung cancer treated with neoadjuvant immune checkpoint inhibitors. EBioMedicine 2022;84:104265. [Crossref] [PubMed]
- Yang H, Ma W, Sun B, et al. Smoking signature is superior to programmed death-ligand 1 expression in predicting pathological response to neoadjuvant immunotherapy in lung cancer patients. Transl Lung Cancer Res 2021;10:3807-22. [Crossref] [PubMed]
- Rahim MK, Okholm TLH, Jones KB, et al. Dynamic CD8(+) T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes. Cell 2023;186:1127-1143.e18. [Crossref] [PubMed]
- Deng H, Zhou J, Chen H, et al. Impact of lymphadenectomy extent on immunotherapy efficacy in postresectional recurred non-small cell lung cancer: a multi-institutional retrospective cohort study. Int J Surg 2024;110:238-52. [Crossref] [PubMed]
- Bousema JE, Dijkgraaf MGW, van der Heijden EHFM, et al. Endosonography With or Without Confirmatory Mediastinoscopy for Resectable Lung Cancer: A Randomized Clinical Trial. J Clin Oncol 2023;41:3805-15. [Crossref] [PubMed]
- Bousema JE, Aarts MJ, Dijkgraaf MGW, et al. Trends in mediastinal nodal staging and its impact on unforeseen N2 and survival in lung cancer. Eur Respir J 2021;57:2001549. [Crossref] [PubMed]
- Rintoul RC, Tournoy KG, El Daly H, et al. EBUS-TBNA for the clarification of PET positive intra-thoracic lymph nodes-an international multi-centre experience. J Thorac Oncol 2009;4:44-8. [Crossref] [PubMed]
- Ong P, Grosu H, Eapen GA, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for systematic nodal staging of lung cancer in patients with N0 disease by computed tomography and integrated positron emission tomography-computed tomography. Ann Am Thorac Soc 2015;12:415-9. [Crossref] [PubMed]
- Gonzalez MM, Polo CG, Domínguez CE, et al. Negative predictive value of EBUS-TBNA in mediastinal staging of lung cancer. European Respiratory Journal 2020;56:3477.
- Schmidt-Hansen M, Baldwin DR, Hasler E, et al. PET-CT for assessing mediastinal lymph node involvement in patients with suspected resectable non-small cell lung cancer. Cochrane Database Syst Rev 2014;2014:CD009519. [Crossref] [PubMed]
- Nomori H, Watanabe K, Ohtsuka T, et al. The size of metastatic foci and lymph nodes yielding false-negative and false-positive lymph node staging with positron emission tomography in patients with lung cancer. J Thorac Cardiovasc Surg 2004;127:1087-92. [Crossref] [PubMed]
- Defranchi SA, Cassivi SD, Nichols FC, et al. N2 disease in T1 non-small cell lung cancer. Ann Thorac Surg 2009;88:924-8. [Crossref] [PubMed]
- Hu Y, Wang P, Dai W. Heterogeneous uptake of (18)F-FDG and (18)F-PSMA-1007 PET/CT in lung cancer and lymph node metastasis. BMC Pulm Med 2023;23:73. [Crossref] [PubMed]
- Wei B, Jin X, Lu G, et al. A novel nomogram to predict lymph node metastasis in cT1 non-small-cell lung cancer based on PET/CT and peripheral blood cell parameters. BMC Pulm Med 2023;23:44. [Crossref] [PubMed]
- Al Bakir M, Huebner A, Martínez-Ruiz C, et al. The evolution of non-small cell lung cancer metastases in TRACERx. Nature 2023;616:534-42. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Zhang JT, Liu SY, Gao W, et al. Longitudinal Undetectable Molecular Residual Disease Defines Potentially Cured Population in Localized Non-Small Cell Lung Cancer. Cancer Discov 2022;12:1690-701. [Crossref] [PubMed]
- Chen K, Yang F, Shen H, et al. Individualized tumor-informed circulating tumor DNA analysis for postoperative monitoring of non-small cell lung cancer. Cancer Cell 2023;41:1749-1762.e6. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577-86. [Crossref] [PubMed]
- Abbosh C, Frankell AM, Harrison T, et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 2023;616:553-62. [Crossref] [PubMed]
- Visvanathan K, Fackler MS, Zhang Z, et al. Monitoring of Serum DNA Methylation as an Early Independent Marker of Response and Survival in Metastatic Breast Cancer: TBCRC 005 Prospective Biomarker Study. J Clin Oncol 2017;35:751-8. [Crossref] [PubMed]
- Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019;176:98-112.e14. [Crossref] [PubMed]
- Dong Q, Chen C, Hu Y, et al. Clinical application of molecular residual disease detection by circulation tumor DNA in solid cancers and a comparison of technologies: review article. Cancer Biol Ther 2023;24:2274123. [Crossref] [PubMed]
- Wang B, Wang M, Lin Y, et al. Circulating tumor DNA methylation: a promising clinical tool for cancer diagnosis and management. Clin Chem Lab Med 2024; Epub ahead of print. [Crossref]
- Ooki A, Maleki Z, Tsay JJ, et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non-Small Cell Lung Cancer and Serum DNA. Clin Cancer Res 2017;23:7141-52. [Crossref] [PubMed]
- Mathios D, Johansen JS, Cristiano S, et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun 2021;12:5060. [Crossref] [PubMed]
- Jamal-Hanjani M, Hackshaw A, Ngai Y, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol 2014;12:e1001906. [Crossref] [PubMed]
- Chen K, Zhao H, Shi Y, et al. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin Cancer Res 2019;25:7058-67. [Crossref] [PubMed]
- Dong D, Zhang S, Jiang B, et al. Correlation analysis of MRD positivity in patients with completely resected stage I-IIIA non-small cell lung cancer: a cohort study. Front Oncol 2023;13:1222716. [Crossref] [PubMed]
- Zhang R, Zhang X, Huang Z, et al. Development and validation of a preoperative noninvasive predictive model based on circular tumor DNA for lymph node metastasis in resectable non-small cell lung cancer. Transl Lung Cancer Res 2020;9:722-30. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Drilon A, Sugita H, Sima CS, et al. A prospective study of tumor suppressor gene methylation as a prognostic biomarker in surgically resected stage I to IIIA non-small-cell lung cancers. J Thorac Oncol 2014;9:1272-7. [Crossref] [PubMed]
- Liang P, Peng M, Tao J, et al. Development of a genome atlas for discriminating benign, preinvasive, and invasive lung nodules. MedComm (2020) 2024;5:e644.


