
Transcriptome sequencing analysis reveals the molecular mechanism of sepsis-induced muscle atrophy
Highlight box
Key findings
• Three transcriptional phases identified—post-surgery muscle response occurs in three distinct transcriptional phases within 72 hours: phase I (0–12 hours): inflammatory-immune phase; phase II (24 hours): inflammatory-atrophy phase; phase III (48–72 hours): atrophy phase. Critical time point: the 24-hour mark (phase II) is identified as a pivotal point for muscle atrophy, suggesting its potential importance in therapeutic interventions.
What is known and what is new?
• Sepsis-induced skeletal muscle atrophy is a critical condition characterized by significant physiological and biochemical changes. It negatively impacts clinical outcomes, prolongs hospital stays, and increases mortality rates.
• This study uncovers detailed molecular pathways and physiological changes underlying sepsis-induced skeletal muscle atrophy, offering new insights into its progression. The findings provide a solid basis for developing targeted therapies by identifying key regulatory processes and markers that could be modulated to prevent or reduce muscle atrophy.
What is the implication, and what should change now?
• The study enhances understanding of the molecular mechanisms behind sepsis-induced muscle atrophy and identifies potential molecular targets for its prevention and treatment. These findings lay the groundwork for developing targeted therapies to address this critical condition effectively.
• Future research should prioritize exploring and validating these molecular targets in clinical settings. Efforts should focus on translating these findings into practical therapeutic strategies to mitigate muscle atrophy and improve patient outcomes. The study highlights the urgent need for integrating molecular insights into sepsis management protocols to optimize treatment approaches.
Introduction
Skeletal muscle is the most abundant type of tissue in the human body and plays an important role in life activities. Skeletal muscle allows humans to perform daily physical activities and is necessary to ensure a good quality of life (1-3). Skeletal muscle responds to various signals, including mechanical stimuli, nerve damage, hormones/growth factors, cachexia, and various cytokines (4-6). The cross-sectional area (CSA) of fibers in muscle changes constantly to regulate muscle strength and metabolism. Muscle-specific RING finger protein 1 (MuRF1) plays a central role in the age-related loss of muscle mass and reduced growth capacity by regulating cellular stress (7). Muscles are highly vulnerable to various injuries, such as mechanical damage, ischemia, nerve damage, and other pathogenic conditions, of which, mechanical injuries are the most common (8-11). The loss of skeletal muscle mass and function due to various causes may eventually lead to the limitation of body movement and may even be life threatening.
Skeletal muscle atrophy is caused by various conditions, including systemic malnutrition, advanced age, disuse, and severe diseases (e.g., cachexia, diabetes mellitus, and sepsis) (12-16). It is characterized by the loss of muscle mass, the inhibition of protein synthesis, and the acceleration of muscle fiber degradation. Research on the molecular mechanisms of skeletal muscle atrophy mainly focuses on E3 ubiquitin ligases, including MuRF1 and muscle atrophy F-box (MAFbx). Atrophy has been shown to increase the expression of MuRF1 and MAFbx in multiple models of muscle atrophy (17-20). Increases in the expression of activated transcription factor 4 in skeletal muscle fibers effectively induces muscle fiber atrophy (21,22). Therefore, the molecular mechanisms of skeletal muscle atrophy are very complex.
Sepsis is a life-threatening organ dysfunction arising from a dysregulated host response to an infection (23-25). Patients with severe sepsis can develop muscle atrophy rapidly, especially if sepsis is combined with multiple organ dysfunction. However, there is a lack of standardized cross-sectional and temporal studies on skeletal muscle atrophy (26). Muscle atrophy occurs early and rapidly in the first week in patients with severe sepsis and is more severe in patients with multiple organ failure, which provides insights into skeletal muscle atrophy due to sepsis (27). Klaude et al. (28) studied skeletal muscle protein metabolism and gene expression in septic patients (28). Notably, various inflammatory cytokines activate multiple signaling pathways, including nuclear factor kappa-B (NF-κB), Janus-activated kinase/signal transducer and activator of transcription (JAK/STAT), and p38 mitogen-activated protein kinase (MAPK). Disrupting the balance between protein synthesis and degradation in skeletal muscle can lead to skeletal muscle atrophy (29-31). Sepsis also induces damage to substantial mitochondria in skeletal muscle, mitochondrial dysfunction in skeletal muscle is associated with a significant increase in autophagy, and the induction of autophagy is more severe in exercise muscle (32).
These studies provided relevant gene expression data; however, as their time point design was not well developed, the results of the normalized time points could not be understood. Thus, a more refined time point design was needed to improve study reliability. A more comprehensive analysis was also needed to obtain a more complete gene expression profile of sepsis-induced muscle atrophy in search of more valuable information. Sepsis-induced muscle atrophy may have a significant effect on would healing and the quality of life of patients with sepsis. However, current therapeutic approaches often fail to produce acceptable outcomes. Thus, there is an urgent need to understand the molecular mechanisms of sepsis-induced muscle atrophy to develop effective therapeutic strategies.
Cecal ligation and puncture (CLP)-induced sepsis is considered similar to that in humans, and thus is often chosen as a model for the study of sepsis induced by multiple microorganisms (33). CLP is considered the gold standard model for animal studies of sepsis. Inflammatory responses are induced by inducing infections with mixed bacterial flora and necrotic tissues, and the intestinal flora plays an important role in sepsis (34,35).
When sepsis occurs, proteolysis in the skeletal muscle is elevated during the acute and chronic sepsis phases to promote skeletal muscle atrophy and then recovered in the late sepsis phase (36). This type of muscle atrophy is regulated by different genes (37), but it is not yet known how sepsis activates the atrophic process through molecular mediators and gene expression during different periods of sepsis.
This study aimed to provide a novel perspective on the transcriptional regulation of sepsis-induced skeletal muscle atrophy. We constructed a skeletal muscle atrophy model of sepsis using the CLP in mouse tibialis anterior muscle. We then identified thousands of differentially expressed genes (DEGs) in the tibialis anterior muscle at different times after sciatic nerve transection by transcriptome sequencing. We then conducted a cluster analysis and a bioinformatics analysis to study gene regulation in skeletal muscle during sepsis. We present this article in accordance with the ARRIVE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1665/rc).
Methods
Animal experiments
A total of 60, 8-week-old, male, Institute of Cancer Research (ICR) mice, weighing 25–30 g, were provided by the Institutional Laboratory Animal Center (Nantong, China) and kept at 22 ℃ under a 12-hour light/dark cycle with free access to food and water. The animal experiments were ethically approved by the Ethics Committee of the Affiliated Hospital of Nantong University (No. 2018-L062), in compliance with the Nantong University’s guidelines for the care and use of animals. A protocol was prepared before the study without registration. The animals were randomly divided into four experimental groups (12, 24, 48, and 72 hours), a sham-operated group and a normal group (n=3 per group).
The mice in the experimental groups were anesthetized with an intraperitoneal injection of pentobarbital (50 mg/kg). A midline dissection was then performed to ligate the distal 2/3 of the cecum, and a 21-gauge needle was used to pierce the ligated portion to express a small amount of feces. The cecum was placed into the abdominal cavity and the incision was sutured. During surgery, the mice were given buprenorphine (0.2 mg/kg subcutaneously) to relieve postoperative pain. Normal saline (50 mg/kg) was injected subcutaneously for postoperative resuscitation. The mice in the sham-operated group were subjected to dissection only, and the cecum was searched out and put back in place.
Muscle mass measurement
The mass of the tibialis anterior muscle is considered an indicator for evaluating muscle mass loss. At the baseline and 12, 24, 48, and 72 hours after CLP, the skin of the mouse hind limbs was peeled off to collect and weigh the tibialis anterior muscle.
Muscle histological changes
The mice were euthanized at four time points (12, 24, 48, and 72 hours) after CLP, and the tibialis anterior muscle samples were dissected, fixed in 4% paraformaldehyde at room temperature overnight, dehydrated in 10–30% sucrose gradients for 3 days, embedded in optimal cutting temperature (OCT) compound and cut into 12-µm-thick sections. Subsequently, the sections were dyed with Cy3 and morphologically observed under Zeiss light microscopy (Axio Imager 2, Jena, Germany). The CSA of the muscle fibers was recorded to evaluate changes in the size of the muscle fibers over time.
Transcriptome sequencing analysis
All the mice were euthanized at four postoperative time points (12, 24, 48, and 72 hours) in the experimental and sham-operated groups, and the tibialis anterior muscle was dissected and stored at −80 ℃ under rapid freezing in liquid nitrogen. Total RNA was extracted from the muscle samples using the mirVana™ miRNA ISOlation kit (Ambion-1561, Texas, USA), and the messenger RNAs (mRNAs) were enriched with magnetic beads carrying Oligo (dT) Recharger (Shanghai Huzheng Company, Shanghai, China) after the deoxyribonuclease digestion of DNA. The mRNAs were broken into short fragments using an interruption reagent. The fragmented mRNA was used as the template, and the first complementary DNA (cDNA) strand was synthesized using six random base primers. The second-strand synthesis reaction system (Kl-131005-30, Shanghai Klang Company, Shanghai, China) was then used to synthesize the second-strand cDNA, and a kit (AM1756, Thermo Fisher, Waltham, USA) was used to purify the double-strand cDNA. The purified double-strand cDNA was end-repaired, A-tailed, and connected to an adapter, and DNA fragment size selection and polymerase chain reaction (PCR) amplification were then performed. The cDNA library was quality checked with an Agilent 2100 Bioanalyzer (City of Santa Clara, Germany) and sequenced using an Illumina HiSeqTM 2500 sequencer (San Diego, USA).
DEG screening
The number of reads mapped to genes in each sample was obtained by htseq count software(https://htseq.readthedocs.io/en/release_0.11.1/count.html). The data were normalized using the estimateSizeFactors function of the DESeq [2012] R package software(https://bioconductor.org). The P value and fold change (FC) value were calculated using the nbinomTest function. The genes with P values <0.05 and FC values >2 were defined as DEGs.
Principal component analysis (PCA) and clustering analysis of DEGs
A PCA was performed based on the quantitative results of the genes to examine the distribution of the samples, explore the relationship between the samples, and validate the experimental design. A closer sample clustering distance or PCA distance indicated a higher similarity between samples. Unsupervised hierarchical clustering of DEGs was also performed. The distance between two pairs of multiple samples was calculated to form a distance matrix. The two closest pairs of classes were merged to form a new class, the distance between the new class and the current class was calculated, and the next closest pairs of classes were then merged until there was only one class. The direct correlations of the samples were calculated based on the expression of the protein-coding DEGs. Generally, the same class of samples appeared in the same cluster by clustering, and the protein-coding DEGs clustered in the same cluster had similar biological functions.
GO and KEGG enrichment analyses of DEGs
After the identification of the DEGs, a Gene Ontology (GO) enrichment analysis of the DEGs was performed to describe their functions based on the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov). The number of DEGs contained in each GO item was counted, and the significance of DEG enrichment in each GO item was calculated using a hypergeometric distribution test. The P values indicated the enrichment significance, with small P values indicating the enrichment of the DEGs in the GO items. The GO categories with P values <0.05 were considered significantly enriched.
A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the protein-coding DEGs (combined with the KEGG annotation results) was performed, and the significance of the enrichment of the DEGs in each entry was calculated using a hypergeometric distribution test. A small P value indicated the enrichment of the DEGs in the pathway. A pathway analysis of the DEGs was conducted to identify the pathway entries that were enriched for the DEGs and determine which cellular pathways might be associated with alterations in the protein-coding DEGs in different samples.
Statistical analysis
A comparative analysis was performed of the RNA-sequencing data to determine whether the same DEGs overlapped in two samples. The following two criteria were applied: (I) the FC value (i.e., the number of FCs in the expression level of the same gene in two samples); and (II) the P value or false discovery rate (FDR) (an adjusted P value). To calculate the FDR value, we first calculated the P value for each gene and then adjusted the P value using the FDR error control method. The default conditions for screening were a P value <0.05 and a FC value >2.
Results
Sepsis-induced skeletal muscle atrophy
The mortality of the mice was assessed during a 72-hour follow-up period. Systemic inflammation during sepsis can have negative effects on peripheral nerves, skeletal muscles, and diaphragm. To assess the effect of sepsis on the skeletal muscle, the mass and CSA of the tibialis anterior muscle were analyzed in a mouse model of CLP (Figure 1). A significant decrease in the mass of the tibialis anterior muscle was observed. In addition, Laminin immunofluorescence staining after CSA measurements was used to assess pathological changes in response to muscle atrophy. The tibialis anterior muscle of the septic mice exhibited thinning and disorganization of muscle fibers and had a reduced CSA. These data suggested that skeletal muscle atrophy occurred in the septic mice.

PCA and DEGs after CLP
A transcriptome sequencing analysis was performed on the RNA samples extracted from the mouse tibialis anterior muscle at 12, 24, 48, and 72 hours after CLP. The PCA analysis results showed that the time after CLP could be divided into three distinct transcriptional phases by converting the two-time nodes (12 and 24 hours) as boundaries between the phases. The clustering analysis provided further evidence of the existence of three transcriptional phases. A large number of DEGs existed in the experimental group after CLP compared to the control group (Figure 2). There were 1,006, 2,044, 1,411, and 1,516 upregulated DEGs, and 1,441, 2,306, 1,950, and 1,644 downregulated DEGs at 12, 24, 48, and 72 hours after CLP, respectively. Notably, there was a significant increase in the number of DEGs at 24 hours after CLP, which suggests that this may be a critical time point for skeletal muscle atrophy.

GO biological process (BP) analysis
A GO analysis was conducted to identify the BPs associated with the DEGs. We identified the enriched BP categories of the upregulated genes at different times after CLP. In transcriptional stage I (Figure 3), the enriched GO BPs included immune system process, response to bacterium, inflammatory response, neutrophil chemotaxis, positive regulation of gene expression, cellular response to lipopolysaccharide, positive regulation of transcription by RNA polymerases II, and positive regulation of apoptotic response, which suggests that the inflammatory response process is mainly activated in this stage. In transcriptional stage I (Figure 4), the enriched GO BPs included positive regulation of ubiquitin-dependent protein catabolic process, proteasomal ubiquitin-independent protein catabolic process, proteasomal-mediated ubiquitin-dependent protein catabolic process, RNA processing, positive regulation of transcription by RNA polymerase II, RNA splicing, ribosome biogenesis, immune system process, and cellular response to lipopolysaccharide, which suggests that proteolysis is extensively activated and proteasomal muscular atrophy is initiated in this stage. In transcriptional stage I (Figure 5), the enriched GO BPs included proteasomal ubiquitin-independent protein catabolic process, proteolysis involving cellular protein catabolic process, proteasomal-mediated ubiquitin-dependent protein catabolic process, ubiquitin-dependent protein catabolic process, ribosomal biogenesis, immune system process, ribosomal RNA processing, response to bacterium, muscle cell development, cellular response to insulin stimulus, and response to denervation involved in regulation of muscle adaptation, which suggests that proteolysis continues in this stage.



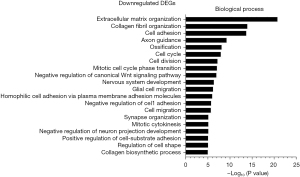
We also identified the BPs enriched in the downregulated genes at different times after CLP. In transcriptional phase I (Figure 6), the enriched BPs included extracellular matrix (ECM) organization, collagen fibril organization, cell adhesion, axon guidance, ossification, cell cycle, cell division, mitotic cell cycle phase transition, negative regulation of canonical Wnt signaling pathway, and nervous system development, which suggests that myoblast growth and metabolism are suppressed during this phase. In transcriptional phase II (Figure 7), the enriched BPs included ECM organization, collagen fibril organization, cell adhesion, nervous system development, negative regulation of canonical Wnt signaling pathways, Wnt signaling pathway, axon guidance, multicellular organism development, ossification, and positive regulation of synaptic assembly, which suggests that myoblast growth is further inhibited in this stage. In transcriptional phase III (Figure 8), the enriched BPs included ECM organization, cell adhesion, collagen fibril organization, multicellular organism development, axon guidance, positive regulation of synaptic assembly, nervous system development, Wnt signaling pathway, cell migration, ossification, transmembrane receptor protein tyrosine kinase signaling pathways, and positive regulation of cell-substrate adhesion, which suggests that myoblast growth continues to be restricted in this stage.


KEGG pathway enrichment analysis
A KEGG pathway analysis was conducted to identify the pathways associated with the DEGs. The enriched KEGG pathways of the upregulated genes at different times after CLP were identified. In transcriptional stage I (Figure 9), the enriched pathways included the tumor necrosis factor (TNF) signaling pathway, interleukin (IL)-17 signaling pathway, NOD-like receptor (NLR) signaling pathway, NF-κB signaling pathway, mitophagy-animal, Toll-like receptor signaling pathway, forkhead box protein O1 (FoxO) signaling pathway, apoptosis, autophagy-animal, C-type lectin receptor signaling pathway, Janus kinase/signal transducer and activator of transcription (JAK-STAT) signaling pathway, and cytokine-cytokine receptor interaction, which suggests that inflammation is mainly activated in this stage. In transcriptional phase II (Figure 10), the enriched pathways included proteasome, the IL-17 signaling pathway, the TNF signaling pathway, autophagy-animal, mitophagy-animal, apoptosis, RNA transport, the FoxO signaling pathway, the NF-κB signaling pathway, and the NLR signaling pathway, which suggests that inflammation continues and the proteasome pathway related to muscle atrophy begins to be activated in this stage. In transcriptional phase III (Figure 11), the enriched pathways included proteasome, autophagy-animal, the IL-17 signaling pathway, the NLR signaling pathway, T helper cell 17 (Th17) differentiation, RNA transport, mitophagy-animal, phagosome, the TNF signaling pathway, metabolism, the FoxO signaling pathway, and the peroxisome proliferator activated receptor (PPAR) signaling pathway, which suggests that the proteasome pathway related to muscle atrophy continues to be activated in this stage.



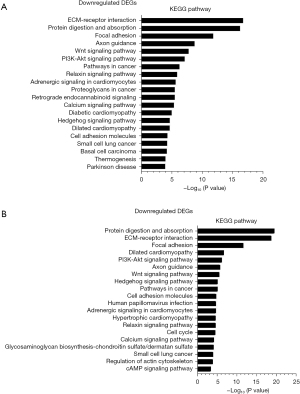
We also examined the enriched KEGG pathways of the downregulated genes at different times after CLP. In transcriptional phase I (Figure 12), the enriched pathways included the ECM-receptor interaction, protein digestion and absorption, axon guidance, focal adhesion, phosphatidylinositol 3-kinase-protein kinase (PI3K-Akt) signaling pathway, cell cycle, cyclic adenosine monophosphate (cAMP) signaling pathway, cell adhesion molecules, gap junction, phagosome, other glycan degradation, apelin signaling pathway, Wnt signaling pathway, and DNA replication pathway, which suggests that myoblast proliferation is inhibited and the inflammatory-immune response is activated in this stage. In transcriptional phase II (Figure 13), the enriched pathways included the ECM-receptor interaction, protein digestion and absorption, focal adhesion, axon guidance, calcium signaling pathway, Wnt signaling pathway, aldosterone synthesis and secretion, PI3K-Akt signaling pathway, adrenergic signaling in cardiomyocytes, proximal tubule bicarbonate reclamation, other glycan degradation, hedgehog signaling pathway, cell cycle, cAMP signaling pathway, cell adhesion molecules, glycine, serine and threonine metabolism, gap junction, and folate biosynthesis, which suggests that inflammation continues and protein synthesis is inhibited in this stage. In transcriptional phase III (Figure 14), the enriched pathways included the ECM-receptor interaction, protein digestion and absorption, focal adhesion, axon guidance, Wnt signaling pathway, PI3K-Akt signaling pathway, relaxin signaling pathway, adrenergic signaling in cardiomyocytes, retrograde endocannabinoid signaling, calcium signaling pathway, hedgehog signaling pathway, cell adhesion molecules, thermogenesis, regulation of actin cytoskeleton, and cAMP signaling pathways, which suggests that the inhibition of protein synthesis is sustained in this stage.


In the three stages of our study, multiple complex networks of gene interactions have been identified. For example, in transcription stage I, there may be a potential interaction between upregulated genes such as TRL 4 and downregulated genes such as Aurkb and Cda 20; Arntl and Cry 2 are up-regulated in transcription stage II, while that of Bub 1 and Aurkb, the down-regulation of Cry 2 and Rb1cc1 and Bub 1 b and Bub 1 may reveal further gene regulatory networks.
Three transcriptional phases
Based on the KEGG and GO analyses, in transcriptional phase I, the inflammatory-immune response is activated and cell proliferation is inhibited, in transcriptional phase II, proteasome is initiated, and in transcriptional phase III, muscle atrophy is extensively activated and protein synthesis is significantly inhibited. Thus, transcriptional stages I–II can be called the “inflammatory-immune stage”, “inflammatory-atrophy stage”, and “atrophy stage”, respectively.
Discussion
The molecular mechanisms underlying sepsis-induced atrophy are unclear. This study conducted a transcriptome sequencing analysis in the time window of sepsis-induced skeletal muscle atrophy and applied various clustering methods and bioinformatics tools to mine the sequencing data to detail the mechanistic information of sepsis-induced skeletal muscle atrophy. The transcriptome sequencing analysis results indicated that a large number of DEGs in the tibialis anterior muscle were up- and downregulated in a time-dependent manner within 12–72 hours after sepsis induction. In total, 1,006, 2,044, 1,411, and 1,516 transcripts were upregulated, and 1,441, 2,306, 1,950, and 1,644 transcripts were downregulated in the tibialis anterior muscle at 12, 24, 48, and 72 hours after CLP, respectively. We also found a significant increase in the number of DEGs 24 hours after sepsis induction, and the GO and KEGG analyses also showed that 24 hours after CLP was a critical time point for muscle gene expression, and most atrophy-related genes were activated at this time point, thus initiating the atrophy process.
In the inflammatory-immune phase, the upregulated genes are related to TNF, IL-17, NLR, NF-κB, Toll-like receptor, FoxO, C-type lectin receptor, JAK-STAT, and cytokine-cytokine receptor interaction (muscle function and homeostasis require cytokine inhibition of AKT activity), which trigger inflammatory responses. Inflammatory pathways may be involved in various types of skeletal muscle atrophy associated with cachexia, aging, and chronic diseases. IL-17 may cause skeletal muscle atrophy by inhibiting myoblast differentiation (38,39). NLRP3 inflammatory vesicles of the NLR family may lead to inflammatory muscle atrophy (40,41). The activation of the NF-κB pathway increases inflammatory muscle damage (42). The increased expression of Toll-like receptors promotes the release of inflammatory factors, thereby triggering skeletal muscle atrophy (43,44). IL-17A-treated myotubes exhibit phosphorylated JAK2/STAT3 signaling and cause skeletal muscle atrophy (45). Lipopolysaccharide induces an increase in NF-κB activity and TNF-α concentration, which in turn affects the protein expression of C2C12 myotubes, leading to skeletal muscle atrophy (46). The motor neuron-specific C-type lectin gene chondrolectin is dysregulated in the mouse model of spinal muscular atrophy (47,48). However, the C-type lectin receptor is a pattern recognition receptor involved in immune and inflammatory responses (49), and its contribution to skeletal muscle atrophy due to inflammation has been less reported. The activation of FoxO1 may trigger sarcopenia (50,51). During sepsis induction, the ubiquitin-proteasome system, autophagy, and FoxO transcription factors in skeletal muscle can lead to skeletal muscle atrophy (42,52). IL-17A promotes skeletal muscle atrophy in lung cancer-induced cachexia through the JAK2/STAT3 pathway (45). In addition to inflammatory signaling pathways, several BPs, including the immune system process, response to bacterium, cellular response to lipopolysaccharides, inflammatory response, viral response, neutrophil chemotaxis, and positive regulation of gene expression, are also activated during the inflammatory phase.
In the inflammatory-atrophy phase, the signaling pathways associated with the upregulated genes include proteasome, IL-17 signaling, TNF signaling, autophagy-animal, mitophagy-animal, apoptosis, RNA transport, the FoxO signaling pathway, NF-κB signaling, and the NLR signaling pathway. The ubiquitin-proteasome system leads to skeletal muscle atrophy through FOXO induction (53). Inflammation-related signaling pathways, such as IL-17 signaling and TNF, are involved in skeletal muscle atrophy. In addition to the signaling pathways involved in atrophy, the ubiquitin-dependent protein catabolic process, proteasomal ubiquitin-independent protein catabolic process, and proteasome-mediated ubiquitin-dependent protein catabolic process are also activated in this stage, which further confirms that this is the inflammatory-atrophy stage.
During the atrophy phase, the signaling pathways related to the upregulated genes include proteasome, autophagy-animal, mitophagy-animal, the IL-17 signaling pathway, protein processing in endoplasmic reticulum, the NLR signaling pathway, Th17 cell differentiation, propanoate metabolism, RNA transport, FoxO, the PPAR signaling pathway, and the TNF signaling pathway. Autophagy and the ubiquitin-proteasome system can cause skeletal muscle atrophy through FOXO induction (53). The Thl 7 cytokine is closely related to muscle injury markers (54). The PPAR signaling pathway directly or indirectly has a major effect on muscle homeostasis (55,56). Phagosomes and mitophagy are essential in the skeletal muscle atrophy phase (57,58). In addition, the following processes are also activated in this stage: proteasomal ubiquitin-independent protein catabolic process, proteolysis involving cellular protein catabolic process, proteasome-mediated ubiquitin-dependent protein catabolic process, ubiquitin-dependent protein catabolic process, response to CLP involved in regulation of muscle adaptation, muscle organ development, ribosomal RNA processing, and cellular response to insulin stimulus.
Numerous DEGs were observed in the skeletal muscle after CLP. The DEGs were mainly involved in the inflammatory response, proteolysis, apoptosis, and autophagy. At 0 to 12 hours after CLP, which is called the inflammatory-immune stage, various inflammations and immune mediators are extensively activated, which is the initiation of skeletal muscle atrophy. It can be said that extensive inflammation initiates skeletal muscle atrophy. At 12 to 24 hours after CLP, which is called the inflammatory-atrophy phase, proteasome begins to be activated and protein catabolism is enhanced, triggering skeletal muscle atrophy. At 48 to 72 hours after CLP, which is the classical skeletal muscle atrophy phase, inflammatory activity largely disappears, and is followed by the extensive activation of proteasomes and the further exacerbation of skeletal muscle catabolism. The findings of the present study not only enrich understandings of the molecular regulatory mechanisms of sepsis-induced muscle atrophy but also provide potential molecular targets for the prevention and treatment of sepsis-induced muscle atrophy and could contribute to the development of targeted therapeutic approaches.
In our study, we have found that in the three stages, multiple complex networks of gene interactions have been identified. In order to further clarify the relationship between these genes, we plan in the subsequent research, using gene coexpression network analysis [such as weighted gene co-expression network analysis (WGCNA)] and protein-protein interaction (PPI) network construction method, system analysis of key genes at different time points, and explore their function and interaction in the research background.
Conclusions
The results of this research demonstrated that tibialis anterior muscle atrophy exacerbated with time after CLP and was accompanied by the altered expression of a large number of genes. The expression profiling analysis showed that there were three transcriptional phases within 72 hours of surgery; that is, transcriptional phase I (0–12 hours), transcriptional phase II (24 hours), and transcriptional phase III (48–72 hours), of which 24 hours may be the critical time point for muscle atrophy. The GO and KEGG analyses showed that the upregulated genes are mainly involved in inflammatory immunity, proteolysis, apoptosis and autophagy, while the downregulated genes are mainly involved in cell proliferation and protein synthesis. We defined these three transcriptional phases as the inflammatory-immune phase, inflammatory-atrophy phase and atrophy phase, respectively. These findings not only enrich understandings of the molecular mechanism of sepsis-induced skeletal muscle atrophy, but also provide a scientific basis for its targeted therapy.
Acknowledgments
The authors would like to thank the Affiliated Hospital of Nantong University for its expert technical assistance and statistical analysis support.
Funding: This research received funding from
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1665/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1665/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1665/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1665/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiments were ethically approved by the Ethics Committee of the Affiliated Hospital of Nantong University (No. 2018-L062), in compliance with the Nantong University’s guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Su X, Shen Y, Kim IM, et al. Extracellular Vesicles for Muscle Atrophy Treatment. Adv Exp Med Biol 2023;1418:119-26. [Crossref] [PubMed]
- Ji Y, Li M, Chang M, et al. Inflammation: Roles in Skeletal Muscle Atrophy. Antioxidants (Basel) 2022;11:1686. [Crossref] [PubMed]
- Huang L, Li M, Deng C, et al. Potential Therapeutic Strategies for Skeletal Muscle Atrophy. Antioxidants (Basel) 2022;12:44. [Crossref] [PubMed]
- Khalil R. Ubiquitin-Proteasome Pathway and Muscle Atrophy. Adv Exp Med Biol 2018;1088:235-48. [Crossref] [PubMed]
- Barreiro E, Jaitovich A. Muscle atrophy in chronic obstructive pulmonary disease: molecular basis and potential therapeutic targets. J Thorac Dis 2018;10:S1415-24. [Crossref] [PubMed]
- Frost RA, Nystrom GJ, Jefferson LS, et al. Hormone, cytokine, and nutritional regulation of sepsis-induced increases in atrogin-1 and MuRF1 in skeletal muscle. Am J Physiol Endocrinol Metab 2007;292:E501-12. [Crossref] [PubMed]
- Hwee DT, Baehr LM, Philp A, et al. Maintenance of muscle mass and load-induced growth in Muscle RING Finger 1 null mice with age. Aging Cell 2014;13:92-101. [Crossref] [PubMed]
- Huard J, Li Y, Fu FH. Muscle injuries and repair: current trends in research. J Bone Joint Surg Am 2002;84:822-32.
- Zhang L, Li M, Wang W, et al. Celecoxib alleviates denervation-induced muscle atrophy by suppressing inflammation and oxidative stress and improving microcirculation. Biochem Pharmacol 2022;203:115186. [Crossref] [PubMed]
- Sun J, Yang H, Yang X, et al. Global alternative splicing landscape of skeletal muscle atrophy induced by hindlimb unloading. Ann Transl Med 2021;9:643. [Crossref] [PubMed]
- Huang Z, Zhong L, Zhu J, et al. Inhibition of IL-6/JAK/STAT3 pathway rescues denervation-induced skeletal muscle atrophy. Ann Transl Med 2020;8:1681. [Crossref] [PubMed]
- Dolly A, Dumas JF, Servais S. Cancer cachexia and skeletal muscle atrophy in clinical studies: what do we really know? J Cachexia Sarcopenia Muscle 2020;11:1413-28. [Crossref] [PubMed]
- Park MJ, Choi KM. Interplay of skeletal muscle and adipose tissue: sarcopenic obesity. Metabolism 2023;144:155577. [Crossref] [PubMed]
- Haberecht-Müller S, Krüger E, Fielitz J. Out of Control: The Role of the Ubiquitin Proteasome System in Skeletal Muscle during Inflammation. Biomolecules 2021;11:1327. [Crossref] [PubMed]
- Wu R, Jiang H, Mao G, et al. Sepsis prognosis related scoring standards: a comprehensive review. Biotarget 2022;5:3.
- Ma W, Zhang R, Huang Z, et al. PQQ ameliorates skeletal muscle atrophy, mitophagy and fiber type transition induced by denervation via inhibition of the inflammatory signaling pathways. Ann Transl Med 2019;7:440. [Crossref] [PubMed]
- Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab 2014;307:E469-84. [Crossref] [PubMed]
- Shen Y, Li M, Wang K, et al. Diabetic Muscular Atrophy: Molecular Mechanisms and Promising Therapies. Front Endocrinol (Lausanne) 2022;13:917113. [Crossref] [PubMed]
- Chen X, Li M, Chen B, et al. Transcriptome sequencing and analysis reveals the molecular mechanism of skeletal muscle atrophy induced by denervation. Ann Transl Med 2021;9:697. [Crossref] [PubMed]
- Ma W, Cai Y, Shen Y, et al. HDAC4 Knockdown Alleviates Denervation-Induced Muscle Atrophy by Inhibiting Myogenin-Dependent Atrogene Activation. Front Cell Neurosci 2021;15:663384. [Crossref] [PubMed]
- Ebert SM, Dyle MC, Bullard SA, et al. Identification and Small Molecule Inhibition of an Activating Transcription Factor 4 (ATF4)-dependent Pathway to Age-related Skeletal Muscle Weakness and Atrophy. J Biol Chem 2015;290:25497-511. [Crossref] [PubMed]
- Ebert SM, Monteys AM, Fox DK, et al. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol 2010;24:790-9. [Crossref] [PubMed]
- Shankar-Hari M, Phillips GS, Levy ML, et al. Developing a New Definition and Assessing New Clinical Criteria for Septic Shock: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:775-87. [Crossref] [PubMed]
- Wu C, Chen Y, Zhou P, et al. Recombinant human angiotensin-converting enzyme 2 plays a protective role in mice with sepsis-induced cardiac dysfunction through multiple signaling pathways dependent on converting angiotensin II to angiotensin 1-7. Ann Transl Med 2023;11:13. [Crossref] [PubMed]
- Gou T, Jin X, Xia J. Idebenone reduces sepsis-induced oxidative stress and apoptosis in hepatocytes via RAGE/p38 signaling. Ann Transl Med 2022;10:1363. [Crossref] [PubMed]
- Churpek MM, Zadravecz FJ, Winslow C, et al. Incidence and Prognostic Value of the Systemic Inflammatory Response Syndrome and Organ Dysfunctions in Ward Patients. Am J Respir Crit Care Med 2015;192:958-64. [Crossref] [PubMed]
- Puthucheary ZA, Rawal J, McPhail M, et al. Acute skeletal muscle wasting in critical illness. JAMA 2013;310:1591-600. [Crossref] [PubMed]
- Klaude M, Mori M, Tjäder I, et al. Protein metabolism and gene expression in skeletal muscle of critically ill patients with sepsis. Clin Sci (Lond) 2012;122:133-42. [Crossref] [PubMed]
- Huang Z, Zhu J, Sun J, et al. Effect of mammalian target of rapamycin signaling pathway on nerve regeneration. Biotarget 2018;2:18.
- Thoma A, Lightfoot AP. NF-kB and Inflammatory Cytokine Signalling: Role in Skeletal Muscle Atrophy. Adv Exp Med Biol 2018;1088:267-79. [Crossref] [PubMed]
- Webster JM, Kempen LJAP, Hardy RS, et al. Inflammation and Skeletal Muscle Wasting During Cachexia. Front Physiol 2020;11:597675. [Crossref] [PubMed]
- Stana F, Vujovic M, Mayaki D, et al. Differential Regulation of the Autophagy and Proteasome Pathways in Skeletal Muscles in Sepsis. Crit Care Med 2017;45:e971-9. [Crossref] [PubMed]
- Dejager L, Pinheiro I, Dejonckheere E, et al. Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol 2011;19:198-208. [Crossref] [PubMed]
- Mattick JS, Yang Q, Orman MA, et al. Long-term gene expression profile dynamics following cecal ligation and puncture in the rat. J Surg Res 2012;178:431-42. [Crossref] [PubMed]
- Gong S, Yan Z, Liu Z, et al. Intestinal Microbiota Mediates the Susceptibility to Polymicrobial Sepsis-Induced Liver Injury by Granisetron Generation in Mice. Hepatology 2019;69:1751-67. [Crossref] [PubMed]
- Voisin L, Breuillé D, Combaret L, et al. Muscle wasting in a rat model of long-lasting sepsis results from the activation of lysosomal, Ca2+ -activated, and ubiquitin-proteasome proteolytic pathways. J Clin Invest 1996;97:1610-7. [Crossref] [PubMed]
- Hasselgren PO, Menconi MJ, Fareed MU, et al. Novel aspects on the regulation of muscle wasting in sepsis. Int J Biochem Cell Biol 2005;37:2156-68. [Crossref] [PubMed]
- Beringer A, Miossec P. Systemic effects of IL-17 in inflammatory arthritis. Nat Rev Rheumatol 2019;15:491-501. [Crossref] [PubMed]
- Obradović H, Krstić J, Kukolj T, et al. Doxycycline Inhibits IL-17-Stimulated MMP-9 Expression by Downregulating ERK1/2 Activation: Implications in Myogenic Differentiation. Mediators Inflamm 2016;2016:2939658. [Crossref] [PubMed]
- Yin X, Han GC, Jiang XW, et al. Increased Expression of the NOD-like Receptor Family, Pyrin Domain Containing 3 Inflammasome in Dermatomyositis and Polymyositis is a Potential Contributor to Their Pathogenesis. Chin Med J (Engl) 2016;129:1047-52. [Crossref] [PubMed]
- Chang L, Niu F, Chen J, et al. Ghrelin improves muscle function in dystrophin-deficient mdx mice by inhibiting NLRP3 inflammasome activation. Life Sci 2019;232:116654. [Crossref] [PubMed]
- Zhang H, He F, Zhou L, et al. Activation of TLR4 induces inflammatory muscle injury via mTOR and NF-κB pathways in experimental autoimmune myositis mice. Biochem Biophys Res Commun 2022;603:29-34. Erratum in: Biochem Biophys Res Commun 2022;606:182. [Crossref] [PubMed]
- Yadav A, Dahuja A, Dabur R. Dynamics of Toll-like Receptors Signaling in Skeletal Muscle Atrophy. Curr Med Chem 2021;28:5831-46. [Crossref] [PubMed]
- Bohnert KR, Goli P, Roy A, et al. The Toll-Like Receptor/MyD88/XBP1 Signaling Axis Mediates Skeletal Muscle Wasting during Cancer Cachexia. Mol Cell Biol 2019;39:e00184-19. [Crossref] [PubMed]
- Ying L, Yao Y, Lv H, et al. IL-17A contributes to skeletal muscle atrophy in lung cancer-induced cachexia via JAK2/STAT3 pathway. Am J Physiol Cell Physiol 2022;322:C814-24. [Crossref] [PubMed]
- Fang WY, Tseng YT, Lee TY, et al. Triptolide prevents LPS-induced skeletal muscle atrophy via inhibiting NF-κB/TNF-α and regulating protein synthesis/degradation pathway. Br J Pharmacol 2021;178:2998-3016. [Crossref] [PubMed]
- Zhang Z, Lotti F, Dittmar K, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008;133:585-600. [Crossref] [PubMed]
- Bäumer D, Lee S, Nicholson G, et al. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 2009;5:e1000773. [Crossref] [PubMed]
- Del Fresno C, Iborra S, Saz-Leal P, et al. Flexible Signaling of Myeloid C-Type Lectin Receptors in Immunity and Inflammation. Front Immunol 2018;9:804. [Crossref] [PubMed]
- Kamei Y, Miura S, Suzuki M, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 2004;279:41114-23. [Crossref] [PubMed]
- Sun H, Sun J, Li M, et al. Transcriptome Analysis of Immune Receptor Activation and Energy Metabolism Reduction as the Underlying Mechanisms in Interleukin-6-Induced Skeletal Muscle Atrophy. Front Immunol 2021;12:730070. [Crossref] [PubMed]
- Parveen A, Bohnert KR, Tomaz da Silva M, et al. MyD88-mediated signaling intercedes in neurogenic muscle atrophy through multiple mechanisms. FASEB J 2021;35:e21821. [Crossref] [PubMed]
- Milan G, Romanello V, Pescatore F, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun 2015;6:6670. [Crossref] [PubMed]
- Sugama K, Suzuki K, Yoshitani K, et al. IL-17, neutrophil activation and muscle damage following endurance exercise. Exerc Immunol Rev 2012;18:116-27.
- Manickam R, Duszka K, Wahli W. PPARs and Microbiota in Skeletal Muscle Health and Wasting. Int J Mol Sci 2020;21:8056. [Crossref] [PubMed]
- Yan Y, Li M, Lin J, et al. Adenosine monophosphate activated protein kinase contributes to skeletal muscle health through the control of mitochondrial function. Front Pharmacol 2022;13:947387. [Crossref] [PubMed]
- Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001;294:1704-8. [Crossref] [PubMed]
- Wang W, Shen D, Zhang L, et al. SKP-SC-EVs Mitigate Denervated Muscle Atrophy by Inhibiting Oxidative Stress and Inflammation and Improving Microcirculation. Antioxidants (Basel) 2021;11:66. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


