
A two-sample mendelian randomization study of non-tuberculous mycobacteria infection and lung cancer
Highlight box
Key findings
• The results do not support a genetically predicted causal relationship between non-tuberculous mycobacteria (NTM) and atypical mycobacterial lung infection and lung cancer.
• The association previously detected between the two entities may stem from the influence of confounding variables.
What is known and what is new?
• Non-tuberculous mycobacterial lung disease may coexist or precede lung cancer, yet a causal link remains unproven.
• The findings do not support a genetically predicted causal relationship between NTM and atypical mycobacterial lung infection and lung cancer.
What is the implication, and what should change now?
• The results of this study may provide evidence on the prediction of lung cancer in patients with NTM/atypical mycobacterial lung infection.
• Mendelian randomization analyses based on larger-scale genome-wide association studies summary data and more genetic instruments are needed to validate the results of this study.
Introduction
Lung cancer persists as one of the most predominant causes of cancer-related mortality worldwide (1). According to the GLOBOCAN 2020 estimates, there were approximately 2.21 million new lung cancer cases, accounting for 11.4% of all cancer cases globally and about 1.80 million deaths, representing 18.0% of cancer-related deaths worldwide (2). The World Health Organization (WHO) classification of lung cancer, taking into account the histological basis, acknowledges a myriad of lung cancer subtypes, encompassing epidermoid carcinomas, adenocarcinomas, small cell lung carcinomas (SCLCs), large cell carcinomas, large cell neuroendocrine carcinomas, adenosquamous carcinomas, sarcomatoid carcinomas, and pleomorphic carcinomas, along with numerous additional forms (3). Of critical importance, reports on non-small cell lung carcinomas (NSCLCs) are deemed inadequate unless accompanied by a comprehensive immunohistochemical profiling (3). However, even with the integration of immunohistochemical analysis, differentiating NSCLC subtypes and identifying specific genetic drivers remain challenging, revealing the limitations of current diagnostic approaches. To address this gap, the present study explores the genetic link between non-tuberculous mycobacteria (NTM) infection and lung cancer, aiming to provide a new perspective for improving early lung cancer diagnosis and expanding current diagnostic strategies beyond conventional methods.
The incidence of non-tuberculous mycobacterial disease has been increasing worldwide (4). NTM lung disease is conventionally characterized by its slow progression in non-immunosuppressed patients, with profound implications for their perceived health and prognosis (5,6). NTM lung disease may occur in patients with structural lung diseases such as bronchiectasis or chronic obstructive pulmonary disease (COPD), and structural lung diseases are also related to the development of lung cancer, whether due to chronic inflammation or common risk factors (7-10). Therefore, it suggests that patients with NTM lung infection may also develop pulmonary malignancy, although it may be difficult to diagnose. Previous studies have reported the co-incidence of NTM lung disease and lung cancer (11,12). Another retrospective study showed that chemotherapy given to patients with NTM lung disease can lead to the progression of NTM lung disease if no anti-NTM treatment is used (13). Results from Kusumoto et al. using both prospective and retrospective methods showed that the incidence of lung cancer in NTM lung disease patients was 124.6 per 100,000 person-years, which was higher than that in Japan (10). However, the causal association between NTM infection and the development of lung cancer has never been reported.
Mendelian randomization (MR) constitutes an instrumental variable (IV) technique that exploits single nucleotide polymorphisms (SNPs) as endogenous instruments to deduce causative associations between phenotypic traits, thereby inherently mitigating biases emanating from confounding variables and the issue of reverse causality (14-17). A two-sample MR design framework was implemented to determine the relationship between NTM/atypical mycobacterial lung infection and lung cancer. The results of this study may provide evidence on the prediction of lung cancer in patients with NTM/atypical mycobacterial lung infection. We present this article in accordance with the STROBE-MR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1268/rc).
Methods
Study design
This article is an MR study. The data for this study were obtained from publicly available databases and published literature data and did not require ethical approval and written informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Figure 1 briefly describes the MR design between NTM and atypical mycobacterial lung infection and lung cancer.
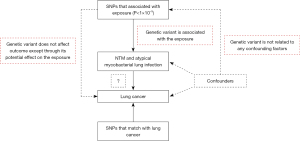
MR is an analytical method based on genetic IVs that aims to infer a causal relationship between an exposure factor (e.g., disease or environmental factors) and an outcome (e.g., lung cancer) through genetic variations (typically SNPs). The principle of MR is similar to the principle of randomized controlled trials in that the distribution of individual genetic variation is random at birth and is not affected by external circumstances or behaviors, thus reducing the effects of confounders and reverse causality (16,18,19). This study used MR methods to assess the genetic causal relationship between NTM infection and lung cancer. Genetic variants that reflect NTM infection were selected as IVs to explore the association of these variants with lung cancer. IVs are at the heart of MR analysis and are used to indirectly measure the impact of exposure factors on outcomes. In order for the IVs to be valid, the following three assumptions must be met (already in the Methods section). (I) Relevance: the IVs must be significantly related to the exposure factors (e.g., NTM infection). (II) Independence: the IVs are not associated with confounders in the exposure-outcome relationship. (III) Exclusion restriction: the IVs only affect the outcome through the exposure and do not directly affect the outcome. The present study identified SNPs significantly associated with NTM infection in the GWASs as IVs.
Data source
The sample sizes of NTM infection, atypical mycobacterium lung infection, lung cancer, squamous cell lung carcinoma (SCC), lung adenocarcinoma (LAC), SCLC were 341,610, 213,312, 374,687, 63,053, 66,756, and 24,108, respectively. The genome-wide association studies (GWAS) data for NTM infection were extracted from a cross-population atlas of genetic associations for 220 human phenotypes (20). There were 26 NTM infection cases and 341,584 control cases. The data for atypical mycobacterium lung infection included 141 cases and 213,171 controls. The summary data for lung cancer were obtained from the UK Biobank (https://gwas.mrcieu.ac.uk/datasets/ieu-b-4955/). There were 2,671 lung cancer cases and 372,016 control. Details on data sources are listed in Table S1.
IVs
In the present study, the IVs incorporated were subject to stringent selection criteria as follows. (I) Initially, SNPs demonstrating genome-wide significant association with NTM infections and atypical mycobacterial pulmonary infections were screened, meeting the P<1×10−5 threshold. (II) SNPs with a minor allele frequency (MAF) >0.01 were selected. (III) SNP linkage disequilibrium (LD) was addressed by excluding SNPs based on the criterion of R2<0.001, employing a window size of 10,000 kb (21). (IV) When an identified IV was absent in the summary data for the outcome, high-LD proxies (R2>0.8) were sought and used as surrogate SNPs. (V) The F statistic for each SNP within the IV was calculated to evaluate instrument strength, aiming to mitigate potential weak instrument bias between the IV and the exposure factor. The formula for computing F is F = R2 * (N − 2)/(1 − R2), where R2 denotes the proportion of variance in the exposure explained by the SNP within the IV. The F statistic was required to be higher than 10 for inclusion (22).
Statistical analysis
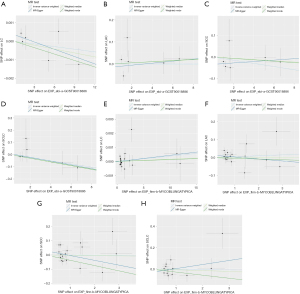
In this analysis, the inverse variance weighted (IVW) approach was the main MR analytical method to assess the causal relationship between the NTM infection and the risk of lung cancer using odds ratios (ORs) and their corresponding 95% confidence intervals (CIs). IVW computes a weighted average of effect sizes, assigning weights proportional to the inverse variance of each SNP (23). To ensure the robustness of the IVW findings, alternative MR approaches such as MR-Egger (24), weighted median (25), and weight mode (26) were also employed. The MR-Egger method accounts for the presence of an intercept term, providing unbiased causal effect estimates even in the presence of horizontal pleiotropy. The weighted median method assumes half of the IVs to be valid in estimating the causal association between the exposure and outcome. The weight mode method estimates the causal effect by focusing on the subset harboring the maximum number of SNPs, a process involving the clustering of SNPs into subsets premised upon the resemblance of causal effect. All analyses were conducted using the ‘TwoSampleMR’ package for R version 4.1.2 (27). The visualization tools included scatter plots and sensitivity analysis diagrams. Funnel plots were used to detect horizontal pleiotropy (27).
Sensitivity analyses were employed to probe for potential pleiotropy in the MR study. Cochran’s Q test was used to assess heterogeneity among the IVs. If P>0.05, heterogeneity was regarded as low, indicating that variability among the IV estimates had a negligible impact on the IVW results. In addition, acknowledging the influence of genetic pleiotropy on the estimated association effects, the MR-Egger regression approach was used to explore the presence of horizontal pleiotropy; an intercept close to zero or non-significant in the MR-Egger regression suggested the absence of substantial pleiotropic effects (28). Furthermore, the MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) method was applied to detect and subsequently exclude potential outliers (i.e., SNPs with P<0.05) that might indicate horizontal pleiotropy, thereby correcting for horizontal pleiotropy through the re-estimation of the causal associations following the removal of the outliers (29). The leave-one-out analysis was applied to evaluate the stability of the results (30).
Results
IVs
In the present study, seven IVs related to NTM infections and 19 IVs pertaining to atypical mycobacterial pulmonary infections were identified. The mean F-statistic for these IVs was 21.26087842, with a minimum F-statistic value of 19.7138682260963 and a maximum value of 24.85183831912. Upon conducting MR analyses with lung cancer and its subtypes as outcomes, six SNPs (rs34962321, rs145417806, rs111716653, rs1437769, rs77472687, and rs9981062) associated with atypical mycobacterial pulmonary infections were found to lack corresponding information in the summary data. Similarly, for NTM infections, two SNPs (rs113700470 and rs185405780) did not match the available summary data. Detailed information on these unmatched SNPs can be found in https://cdn.amegroups.cn/static/public/jtd-24-1268-1.xlsx.
MR
The MR findings revealed no statistically significant causal associations between NTM infections and atypical mycobacterial pulmonary infections with the risk of lung cancer and its subtypes, as shown in Table 1, Figure 2, and Figure S1. IVW showed no significant relationships between NTM infection and the risk of lung cancer (OR =1.00, 95% CI: 1.00–1.00, P=0.50) and no significant relationships between atypical mycobacterial lung infection and the risk of lung cancer (OR =1.00, 95% CI: 1.00–1.00, P=0.47). Regarding the subtypes of lung cancer, IVW showed no significant relationships between NTM infection and LAC (OR =1.00, 95% CI: 0.99–1.02, P=0.99) and between atypical mycobacterial lung infection and LAC (OR =1.01, 95% CI: 0.99–1.02, P=0.54); the OR of the relationship between NTM infection and SCC was 0.99 (95% CI: 0.98–1.01, P=0.27) and the OR of the relationship between atypical mycobacterial lung infection and SCC was 1.00 (95% CI: 0.97–1.03, P=0.97). The IVW results showed no causal relationships between NTM infection and SCLC (OR =0.99, 95% CI: 0.97–1.01, P=0.23) and no causal relationships between atypical mycobacterial lung infection and SCLC (OR =1.00, 95% CI: 0.96–1.03, P=0.89). The results from the MR-Egger, weighted median, and weighted mode methods were consistent with the IVW method.
Table 1
| Exposure | Outcome | N. SNPs | Methods | OR (95% CI) | P |
|---|---|---|---|---|---|
| AMLI | LAC | 17 | Inverse variance weighted | 1.01 (0.99–1.02) | 0.54 |
| 17 | MR-Egger | 0.99 (0.95–1.03) | 0.59 | ||
| 17 | Weighted median | 1.00 (0.98–1.02) | 0.99 | ||
| 17 | Weighted mode | 1.00 (0.96–1.03) | 0.83 | ||
| LC | 19 | Inverse variance weighted | 1.00 (1.00–1.00) | 0.47 | |
| 19 | MR-Egger | 1.00 (1.00–1.00) | 0.54 | ||
| 19 | Weighted median | 1.00 (1.00–1.00) | 0.83 | ||
| 19 | Weighted mode | 1.00 (1.00–1.00) | 0.84 | ||
| SCC | 17 | Inverse variance weighted | 1.00 (0.97–1.03) | 0.97 | |
| 17 | MR-Egger | 0.97 (0.9–1.03) | 0.35 | ||
| 17 | Weighted median | 1.00 (0.97–1.03) | 0.88 | ||
| 17 | Weighted mode | 0.96 (0.92–1.01) | 0.17 | ||
| SCLC | 13 | Inverse variance weighted | 1.00 (0.96–1.03) | 0.89 | |
| 13 | MR-Egger | 1.04 (0.95–1.14) | 0.44 | ||
| 13 | Weighted median | 0.99 (0.95–1.03) | 0.72 | ||
| 13 | Weighted mode | 0.97 (0.91–1.04) | 0.39 | ||
| NTMI | LAC | 6 | Inverse variance weighted | 1.00 (0.99–1.02) | 0.99 |
| 6 | MR-Egger | 1.00 (0.98–1.03) | 0.73 | ||
| 6 | Weighted median | 1.00 (0.99–1.02) | 0.74 | ||
| 6 | Weighted mode | 1.00 (0.99–1.02) | 0.70 | ||
| LC | 6 | Inverse variance weighted | 1.00 (1.00–1.00) | 0.50 | |
| 6 | MR-Egger | 1.00 (1.00–1.00) | 0.21 | ||
| 6 | Weighted median | 1.00 (1.00–1.00) | 0.35 | ||
| 6 | Weighted mode | 1.00 (1.00–1.00) | 0.25 | ||
| SCC | 6 | Inverse variance weighted | 0.99 (0.98–1.01) | 0.27 | |
| 6 | MR-Egger | 1.00 (0.98–1.02) | 0.76 | ||
| 6 | Weighted median | 1.00 (0.98–1.02) | 0.75 | ||
| 6 | Weighted mode | 1.00 (0.98–1.01) | 0.70 | ||
| SCLC | 6 | Inverse variance weighted | 0.99 (0.97–1.01) | 0.23 | |
| 6 | MR-Egger | 0.98 (0.95–1.02) | 0.39 | ||
| 6 | Weighted median | 0.99 (0.96–1.01) | 0.24 | ||
| 6 | Weighted mode | 0.99 (0.96–1.01) | 0.31 |
MR, Mendelian randomization; N. SNPs, number of single-nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; AMLI, atypical mycobacterium lung infection; NTMI, non-tuberculous mycobacterial infection; LAC, lung adenocarcinoma; LC, lung cancer; SCC, squamous cell lung carcinoma; SCLC, small cell lung carcinoma.
Sensitivity testing
The MR PRESSO test showed no incidence of potential pleiotropy except for atypical mycobacterial pulmonary infections and SCC. Two outliers were detected when atypical mycobacterial pulmonary infections were considered as the exposure and SCC as the outcome (Table 2). Following excluding these outliers, the results remained similar, i.e., no significant causal relationships were observed between atypical mycobacterial pulmonary infections and the risk of developing SCC. The results from Cochran’s Q test indicated the presence of heterogeneity in the analysis between atypical mycobacterial pulmonary infections and SCC (Q=46.36, P<0.001) (Table 3). The MR-Egger test showed that the results were not affected by horizontal pleiotropy (Table 3). The funnel plots are shown in Figure S2. The leave-one-out analysis indicated that the causal estimates of the primary analysis and the cancer subtype analysis were not driven by any single SNP. The leave-one-out analysis plots are shown in Figure S3.
Table 2
| Exposure | Outcome | Raw | Outlier corrected | Global P | Number of outliers | Distortion P | |||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | ||||||
| AMLI | LAC | 1.01 (0.99–1.02) | 0.55 | NA | NA | 0.13 | 0 | NA | |
| LC | 1.00 (1.00–1.00) | 0.48 | NA | NA | 0.49 | 0 | NA | ||
| SCC | 1.00 (0.97–1.03) | 0.97 | 0.99 (0.96–1.01) | 0.24 | <0.001 | 2 | 0.66 | ||
| SCLC | 1.00 (0.96–1.03) | 0.89 | NA | NA | 0.11 | 0 | NA | ||
| NTMI | LAC | 1.00 (0.99–1.02) | 0.99 | NA | NA | 0.25 | 0 | NA | |
| LC | 1.00 (1.00–1.00) | 0.53 | NA | NA | 0.26 | 0 | NA | ||
| SCC | 0.99 (0.98–1.00) | 0.26 | NA | NA | 0.63 | 0 | NA | ||
| SCLC | 0.99 (0.98–1.00) | 0.07 | NA | NA | 0.96 | 0 | NA | ||
MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; OR, odds ratio; CI, confidence interval; AMLI, atypical mycobacterium lung infection; NTMI, non-tuberculous mycobacterial infection; LAC, lung adenocarcinoma; LC, lung cancer; SCC, squamous cell lung carcinoma; SCLC, small cell lung carcinoma; NA, not applicable.
Table 3
| Exposure | Outcome | Heterogeneity | Pleiotropy | |||
|---|---|---|---|---|---|---|
| Q statistic (IVW) | P value | MR-Egger intercept | P value | |||
| AMLI | LAC | 21.87 | 0.15 | 0.02 | 0.39 | |
| LC | 18.62 | 0.42 | −1.42 | 0.88 | ||
| SCC | 46.36 | <0.001 | 0.03 | 0.31 | ||
| SCLC | 18.64 | 0.10 | −0.03 | 0.37 | ||
| NTMI | LAC | 9.01 | 0.11 | −0.02 | 0.64 | |
| LC | 7.79 | 0.17 | 0.00 | 0.24 | ||
| SCC | 3.85 | 0.57 | −0.03 | 0.22 | ||
| SCLC | 1.32 | 0.93 | 0.01 | 0.84 | ||
MR, Mendelian randomization; IVW, inverse variance weighted; AMLI, atypical mycobacterium lung infection; NTMI, non-tuberculous mycobacterial infection; LAC, lung adenocarcinoma; LC, lung cancer; SCC, squamous cell lung carcinoma; SCLC, small-cell lung carcinoma.
Discussion
It is the first study to investigate the causal relationship between NTM and atypical mycobacterial lung infections and lung cancer using MR. The present two-sample MR analyses did not reveal evidence supporting that NTM or atypical mycobacterial lung infections were associated with lung cancer.
Previous investigations demonstrated that NTM may sometimes be isolated from respiratory specimens in patients with lung cancer (12,31,32). In a retrospective matched cohort study investigating the association of pulmonary NTM with the clinical features and outcomes of patients with lung cancer (32), the prevalence of pulmonary NTM in patients with lung cancer was 2.5% (138/5,418). Another study conducted in Japan showed that the incidence of pulmonary NTM in untreated lung cancer patients was 1.4% (31). The possible reasons for the variation in NTM incidences in lung cancer patients may be the small sample size of the studies and the different ethnicities of the patients. In addition, previous studies have been performed to evaluate the incidence of lung cancer in NTM lung infection, in which 2.4% to 7.3% of patients with NTM lung disease or respiratory cultures growing Mycobacterium avium complex (MAC) had lung cancer (11,12,33). Several potential reasons may explain the relationship between NTM and atypical mycobacterial lung infection and lung cancer in observational studies. Chronic pulmonary inflammation is a compelling etiological element in the pathogenesis of lung cancer, as evidenced by its association with various chronic respiratory conditions, such as idiopathic pulmonary fibrosis, COPD, and tuberculosis (34,35). In particular, neutrophilic inflammation is pivotal in the progression of MAC lung disease (36) and can engender reactive oxygen and nitrogen species, thereby causing genomic aberrations (37). Furthermore, the disease microenvironment in MAC lung disease is characterized by increased levels of pro-inflammatory cytokines, including tumor necrosis factor-α and interleukin-6 (38), which may exert their effects on epithelial cells to transcriptionally upregulate anti-apoptotic gene expression (39).
Albeit with significant findings indicating associations between NTM and atypical mycobacterial lung infection and lung cancer, the results were from observational studies. One of the limitations of previous studies may be that the potential confounding factors were not comprehensively adjusted. This issue could be properly addressed using the MR methods. The results of the present MR study suggest no significant causal associations. The findings of this study indicate that causal evidence on the prediction of lung cancer in patients with NTM/atypical mycobacterial lung infection could be due to common confounders. In future studies, emphasis should be paid to proven risk factors of lung cancer in patients with NTM or atypical mycobacterial lung infection.
However, this study has some limitations. First, the MR analysis was confined to a study population of European ancestry; hence, the generalizability of the findings to broader populations remains unestablished. Second, although the potential overlap of participants between the exposure and outcome datasets is plausible, quantifying the extent of such sample convergence poses a challenge. However, the utilization of highly robust instruments (as exemplified by F-values all >10) in this study should effectively mitigate any potential biases stemming from participant overlap (40). Third, the limited number of SNPs chosen as IVs may account for only a minor portion of the exposure variability, potentially attenuating the statistical power of the derived causal estimates. Based on the limitations, caution should be paid to the interpretation of findings in this study.
Conclusions
The findings of the present two-sample MR analysis did not support a genetically predicted causal relationship between NTM and atypical mycobacterial lung infection and lung cancer. Consequently, the association previously detected in observational studies between the two entities may stem from the influence of confounding variables. MR analyses based on larger-scale GWAS summary data and more genetic instruments are needed to validate the results of this study.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE-MR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1268/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1268/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1268/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This article is a Mendelian randomization study. The data for this study were obtained from publicly available databases and published literature data and did not require ethical approval and written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clin North Am 2019;103:463-73. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- de Sousa VML, Carvalho L. Heterogeneity in Lung Cancer. Pathobiology 2018;85:96-107. [Crossref] [PubMed]
- Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med 2015;36:13-34. [Crossref] [PubMed]
- Asakura T, Funatsu Y, Ishii M, et al. Health-related quality of life is inversely correlated with C-reactive protein and age in Mycobacterium avium complex lung disease: a cross-sectional analysis of 235 patients. Respir Res 2015;16:145. [Crossref] [PubMed]
- Hayashi M, Takayanagi N, Kanauchi T, et al. Prognostic factors of 634 HIV-negative patients with Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 2012;185:575-83. [Crossref] [PubMed]
- Honda JR, Knight V, Chan ED. Pathogenesis and risk factors for nontuberculous mycobacterial lung disease. Clin Chest Med 2015;36:1-11. [Crossref] [PubMed]
- Shu CC, Wu MF, Pan SW, et al. Host immune response against environmental nontuberculous mycobacteria and the risk populations of nontuberculous mycobacterial lung disease. J Formos Med Assoc 2020;119:S13-22. [Crossref] [PubMed]
- Chung WS, Lin CL, Hsu WH, et al. Increased risk of lung cancer among patients with bronchiectasis: a nationwide cohort study. QJM 2016;109:17-25. [Crossref] [PubMed]
- Kusumoto T, Asakura T, Suzuki S, et al. Development of lung cancer in patients with nontuberculous mycobacterial lung disease. Respir Investig 2019;57:157-64. [Crossref] [PubMed]
- Winthrop KL, McNelley E, Kendall B, et al. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. Am J Respir Crit Care Med 2010;182:977-82. [Crossref] [PubMed]
- Lande L, Peterson DD, Gogoi R, et al. Association between pulmonary mycobacterium avium complex infection and lung cancer. J Thorac Oncol 2012;7:1345-51. [Crossref] [PubMed]
- Tsuji T, Tsuyuguchi K, Tachibana K, et al. Analysis of the impact of lung cancer treatment on nontuberculous mycobacterial lung diseases. Respir Investig 2017;55:45-50. [Crossref] [PubMed]
- Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA 2017;318:1925-6. [Crossref] [PubMed]
- Lawlor DA, Harbord RM, Sterne JA, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133-63. [Crossref] [PubMed]
- Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [Crossref] [PubMed]
- Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89-98. [Crossref] [PubMed]
- Burgess S, Davey Smith G, Davies NM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res 2019;4:186. [Crossref] [PubMed]
- Sanderson E, Glymour MM, Holmes MV, et al. Mendelian randomization. Nat Rev Methods Primers 2022;2:6. [Crossref] [PubMed]
- Sakaue S, Kanai M, Tanigawa Y, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet 2021;53:1415-24. [Crossref] [PubMed]
- 1000 Genomes Project Consortium; Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061-73.
- Burgess S, Thompson SGCRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 2011;40:755-64. [Crossref] [PubMed]
- Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658-65. [Crossref] [PubMed]
- Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512-25. [Crossref] [PubMed]
- Bowden J, Davey Smith G, Haycock PC, et al. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol 2016;40:304-14. [Crossref] [PubMed]
- Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017;46:1985-98. [Crossref] [PubMed]
- Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [Crossref] [PubMed]
- Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377-89. [Crossref] [PubMed]
- Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693-8. [Crossref] [PubMed]
- Burgess S, Bowden J, Fall T, et al. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017;28:30-42. [Crossref] [PubMed]
- Tamura A, Hebisawa A, Sagara Y, et al. Pulmonary nontuberculous mycobacteriosis in patients with lung cancer. Kekkaku 2004;79:367-73.
- Liao TY, Wang JY, Shih JY. Association of pulmonary nontuberculous mycobacteria with the outcomes of patients with lung cancer: A retrospective matched cohort study with a special emphasis on the impact of chemotherapy. J Microbiol Immunol Infect 2023;56:392-9. [Crossref] [PubMed]
- Hosoda C, Hagiwara E, Shinohara T, et al. Clinical characteristics of pulmonary Mycobacterium avium complex infection complicated with lung cancer. Kekkaku 2014;89:691-5.
- Liang HY, Li XL, Yu XS, et al. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: a systematic review. Int J Cancer 2009;125:2936-44. [Crossref] [PubMed]
- Chatila WM, Thomashow BM, Minai OA, et al. Comorbidities in chronic obstructive pulmonary disease. Proc Am Thorac Soc 2008;5:549-55. [Crossref] [PubMed]
- Inomata T, Konno S, Nagai K, et al. Neutrophil predominance in bronchoalveolar lavage fluid is associated with disease severity and progression of HRCT findings in pulmonary Mycobacterium avium infection. PLoS One 2018;13:e0190189. [Crossref] [PubMed]
- Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest 2007;117:1175-83. [Crossref] [PubMed]
- Tasaka S, Hasegawa N, Nishimura T, et al. Elevated serum adiponectin level in patients with Mycobacterium avium-intracellulare complex pulmonary disease. Respiration 2010;79:383-7.
- Dheda K, Booth H, Huggett JF, et al. Lung remodeling in pulmonary tuberculosis. J Infect Dis 2005;192:1201-9. [Crossref] [PubMed]
- Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013;178:1177-84. [Crossref] [PubMed]


