Immunoproteasome deficiency in non-small cell lung cancer and its relevance to immunotherapy
A key characteristic of tumors associated with poor prognosis is their ability to escape the rigorous scrutiny of the immune system (1). Understanding how they do so not only advances our knowledge of fundamental mechanisms underlying cancer, but also improves our ability to detect and assess tumors as well as develop and deploy the most effective treatments to remove them. Down-regulation of the class I major histocompatibility (MHC) complex on their surface is a well-known mechanism by which tumor cells attempt to evade detection of their mutational load via the corresponding presented neo-antigens. The immune system reacts to the lack of MHC class I molecules by activating natural killer cells, which destroy these aberrant cells and restore balance. In their recent publication, Tripathi et al. describe an alternative way by which lung cancer cells may evade immune control (2). They have discovered that lung cancer cells that are more mesenchymal-like show a significant down-regulation of the immunoproteasome, compared to those with an epithelial-like phenotype. These cells are also associated with decreased survival and reduced time to progression. This implies that deficiency in the ability to efficiently produce and present immunopeptides, rather than in MHC class I molecules themselves, may make tumor cells more successful in evading immune control. Such a mechanism has been reported previously for e.g., melanoma (3). Consequently, strategies that rectify this deficiency have the potential to enhance emerging immunotherapeutic approaches in non-small cell lung cancer (NSCLC) and beyond (4).
The immunoproteasome is an important, regulated component of the immune system (Figure 1). It is closely related to the 26S proteasome, a multi-protein complex responsible for protein degradation in cells (5). The proteasome core is formed by the 20S subunit, which consists of two identical halves. Each half is made of two rings of seven proteins each (α1–α7 and β1–β7) that together form a four-ring barrel structure. Three subunits of the innermost β rings have proteolytic activity (β1, β2, β5), while the α rings govern accessibility to these proteolytic enzymes. The 20S core maintains cellular homeostasis by converting misfolded or modified proteins into peptides. This process is controlled by the 19S regulatory subunit, which caps either end of the 20S core and recognizes ubiquitinylated proteins destined for degradation. A subset of the peptides the proteasome produces at any time is transported to the endoplasmic reticulum (ER), where they can bind to MHC class I molecules. The resulting complexes then relocate via the Golgi apparatus to the cell surface, where they are exposed. CD8+ T cells bind strongly if their unique T cell receptor (TCR) sequence matches, then activate and release cytotoxins. This antigen presentation process enables the immune system to sense the presence of “neo-antigens”, including those arising from high mutational load and aberrant protein patterns associated with cancer. Protein degradation and cellular homeostasis remains the main functionality of the ubiquitin-proteasome system (UPS), however (5).
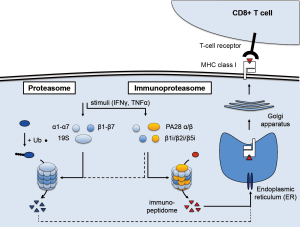
Arising from the proteasome, the immunoproteasome is a dedicated system that can be induced when antigen presentation is of critical importance (6). In situations of pathogenic invasion, e.g., when a virus seeks to replicate and amplify inside infected cells, the expression of three immunoproteasome subunit genes is induced. The corresponding β1i, β2i, and β5i proteins are preferentially incorporated into newly assembled proteasome complexes, where they replace the β1, β2, and β5 subunits. This alters the proteolytic activity and specificity of the proteasome and results in higher yield of peptides. The β5i subunit, also called LMP7 or PSMB8, is particularly important: it is involved in the maturation of the β1i and β2i subunits, and encodes the chymotryptic activity that improves binding to the MHC class I molecules and facilitates T cell recognition. In addition, the 11S complex consisting of the PA28 α and β subunits replaces the 19S regulatory complex, which eliminates the requirement for protein ubiquitinylation. As a result, assembly of the immunoproteasome facilitates protein degradation in general, including proteins of viral origin, and leads to more efficient presentation of the corresponding immunopeptidome to cytotoxic T cells.
A variety of stimuli can induce immunoproteasome expression, most notably interferon-γ (IFNγ). Other inflammatory cytokines such as interferon α and β or tumor necrosis factor α (TNFα), as well as other stimuli such as oxidative stress, also activate the immunoproteasome in a context-dependent way. While additional, non-immune functions have also been reported, the main role of the immunoproteasome is thought to be in immune surveillance (7). Indeed, “professional antigen-presenting cells” such as macrophages, B cells, or dendritic cells, constitutively express the immunoproteasome. More recently, basal expression has also been detected in non-immune cells. Interestingly, this includes epithelial cells of the skin and lung, which also have a higher risk of being exposed to pathogens. Expression of the immunoproteasome at basal levels and the ability to induce strong expression may well reflect this aspect, which coincides with skin and lung tumors typically harboring more non-synonymous mutations than other tumor types (8).
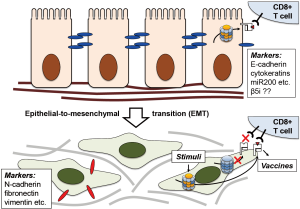
Tripathi et al. are now reporting a previously unappreciated aspect of immunoproteasome regulation: the apparent loss of immunoproteasome expression in NSCLC and its association with a more aggressive and invasive phenotype. While the exact mechanism is currently unknown, the authors illustrate the balance of signal transducer and activator of transcription (STAT) 1 and 3 and the micro-RNA miR-200 that inhibits transforming growth factor β (TGFβ) and therefore STAT3 activation to be key elements in this process. MiR-200 are small, non-coding RNA molecules that function as tumor suppressors by targeting E-cadherin (CDH1) transcriptional repressors (9), thereby preventing loss of E-cadherin expression that is associated with epithelial-to-mesenchymal transition (EMT) (Figure 2). Indeed, the authors show positive correlation of β5i and STAT1, and slight negative correlation of β5i and STAT3 expression, establishing low miR-200, low immunoproteasome, and high STAT3 levels as hallmarks of the mesenchymal phenotype. They also incorporate epigenetic regulation of the immunoproteasome via DNA hypermethylation and the mTOR pathway. Importantly, this enables targeted intervention to restore immunoproteasome expression in mesenchymal cell lines, which the authors achieve by treatment with a demethylating agent 5-aza-2’-deoxycytidine (5-aza-dC) and the mTOR inhibitor rapamycin (2).
The successful recuperation of immunoproteasome expression in lung cancer cells opens an intriguing new avenue to improving T cell-mediated immunotherapy, provided it translates into enhanced immunopeptidome repertoire and corresponding T-cell recognition. The authors show that this might be possible: treatment of mesenchymal cell lines with IFNγ resulted in an increase in the diversity and abundance of presented peptides, and these peptides could trigger cytotoxicity and cell lysis with circulating cytotoxic T-lymphocytes (2). As of yet, the question of how these in vitro experiments might translate into actual in vivo approaches remains unanswered. While a number of possible scenarios can be envisioned, none of them will be straightforward.
The most obvious therapeutic approach would be to simply treat patients with IFNγ to restore immunoproteasome expression. Notably, IFNγ is already FDA-approved for the treatment of chronic granulomatous disease (10) and osteopetrosis, has also been used to treat patients with atopic dermatitis (11) or Crohn’s disease (12), and has been shown to curb infection with Ebola virus (13). In these cases, IFNγ results in the desired enhanced immune sensitivity. However, immune functions beyond antigen presentation are also ascribed to the immunoproteasome, including the regulation of cytokine production via the NF-κB pathway, T cell expansion, and T helper cell differentiation. Moreover, a β5i-specific inhibitor reduces symptoms in several animal models of autoimmune diseases (14). Systemic induction of immunoproteasome expression via IFNγ may therefore not have the desired effect, but could also exacerbate inflammatory conditions. In addition, by triggering stronger activation of professional antigen-presenting cells and cytokine release, it may also cause more extensive cytotoxicity and subsequent depletion. To avoid such systemic responses that hamper therapeutic applications, it may be advisable to implement a targeted delivery approach involving cell surface proteins that are highly expressed on mesenchymal-like cancer cells, e.g., N-cadherin (CDH2).
If immunoproteasome expression and immunopeptide presentation could be restored this way, mounting an effective immune response would also require the corresponding immune cells to efficiently access the tumor site and kill the tumor cells. Tumor-infiltrating lymphocytes widely range in abundance, suggesting that not all tumors will meet this requirement. On the other hand, mesenchymal-like tumor cells are found at the invasive front of tumors or migrating in tissue and generally show decreased adhesion to the extracellular matrix or other cells and increased ability to modulate their environment. They could therefore already be prone to exposure, but simply escape detection due to down-regulated immunoproteasome expression. Restoration of immunoproteasome expression may then be sufficient to unmask them. However, tumor cells often utilize additional, alternative strategies to avoid an immune response, or even use it to their advantage (15). This includes interference with immune checkpoints such as cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) or programmed cell death protein 1 (PD-1). They are expressed on immune cells to prevent tissue damage by curbing the immune response, e.g., via the PD-1 ligand PD-L1 (16). CTLA-4 and PD-1/PD-L1 inhibitors have been FDA-approved for melanoma (17) and have entered clinical trials in lung cancer, yet their efficacy might still benefit from also restoring immunoproteasome function.
An alternative therapeutic avenue that has already shown success in melanoma and other cancer types as well as clinical trials in NSCLC could utilize vaccines. These are often fusion proteins containing a tumor-associated signature and an immune cell activator that is infused or injected, subsequently processed by APCs and presented to naïve T-cells to initiate an immune response in the patient. What makes it particularly intriguing is the possibility of “personalizing” the vaccine based on the specific patterns of each patient (18). For this to be successful, the presented immunopeptides need to be harvested and analyzed for the presence of unusual signatures that could be used to develop vaccines (19). This process still faces significant technical challenges (20), including the relatively large amount of material needed to define such signatures by immunopeptidomics, for which tumor cells need to be collected in sufficient numbers or expanded ex vivo. Harvesting lung epithelial cells is more difficult than skin cells, and the practicability of this approach in treating lung cancer is not yet known. Restoration of immunoproteasome expression ex vivo by IFNγ, 5-aza-dC, or rapamycin to increase immunopeptidome yield may well help to overcome some of these challenges.
While many of these therapeutic aspects will need to be addressed in the long term, the authors’ discovery may have a more immediate impact in the diagnostic field. Notably, their report highlights the value of the histological characterization of tumors based on the expression of the immunoproteasome subunit β5i (PSMB8). The localization of tumor cells with low β5i/PSMB8 expression to its invasive edge may not only help judge how aggressive a tumor is, which other markers such as CDH1/CDH2 may accomplish as well. In addition, it may also help gauge the success of conventional T cell-based immunotherapy vs the need to rely on alternatives such as chimeric antigen receptor T cells (CAR-T cells) (21), based on the likelihood of successful T-cell recognition with or without prior immunoproteasome induction.
Regardless of the success that can ultimately be achieved in any of these aspects, by adding new knowledge Tripathi et al. (2) have already helped expand the therapeutic framework in the rapidly advancing field of cancer immunotherapy, particularly as it applies to lung cancer.
Acknowledgements
The author would like to thank the members of his research group for their helpful input and perspective during the manuscript writing process.
Footnote
Provenance: This is an invited Commentary commissioned by the Section Editor Long Jiang (Second Affiliated Hospital, Institute of Respiratory Diseases, Zhejiang University School of Medicine, Hangzhou, China).
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology 2007;121:1-14. [Crossref] [PubMed]
- Tripathi SC, Peters HL, Taguchi A, et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci U S A 2016;113:E1555-64. [Crossref] [PubMed]
- Dissemond J, Goette P, Moers J, et al. Immunoproteasome subunits LMP2 and LMP7 downregulation in primary malignant melanoma lesions: association with lack of spontaneous regression. Melanoma Res 2003;13:371-7. [Crossref] [PubMed]
- Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288-300. [Crossref] [PubMed]
- Navon A, Ciechanover A. The 26 S proteasome: from basic mechanisms to drug targeting. J Biol Chem 2009;284:33713-8. [Crossref] [PubMed]
- Angeles A, Fung G, Luo H. Immune and non-immune functions of the immunoproteasome. Front Biosci (Landmark Ed) 2012;17:1904-16. [Crossref] [PubMed]
- Kimura H, Caturegli P, Takahashi M, et al. New Insights into the Function of the Immunoproteasome in Immune and Nonimmune Cells. J Immunol Res 2015;2015:541984.
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol 2008;5:115-9. [Crossref] [PubMed]
- Errante PR, Frazão JB, Condino-Neto A. The use of interferon-gamma therapy in chronic granulomatous disease. Recent Pat Antiinfect Drug Discov 2008;3:225-30. [Crossref] [PubMed]
- Brar K, Leung DY. Recent considerations in the use of recombinant interferon gamma for biological therapy of atopic dermatitis. Expert Opin Biol Ther 2016;16:507-14. [Crossref] [PubMed]
- Cui D, Huang G, Yang D, et al. Efficacy and safety of interferon-gamma-targeted therapy in Crohn's disease: a systematic review and meta-analysis of randomized controlled trials. Clin Res Hepatol Gastroenterol 2013;37:507-13. [Crossref] [PubMed]
- Rhein BA, Powers LS, Rogers K, et al. Interferon-γ Inhibits Ebola Virus Infection. PLoS Pathog 2015;11:e1005263. [Crossref] [PubMed]
- Basler M, Mundt S, Bitzer A, et al. The immunoproteasome: a novel drug target for autoimmune diseases. Clin Exp Rheumatol 2015;33:S74-9. [PubMed]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011;331:1565-70. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Boutros C, Tarhini A, Routier E, et al. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat Rev Clin Oncol 2016;13:473-86. [Crossref] [PubMed]
- Cobbold M, De La Peña H, Norris A, et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med 2013;5:203ra125. [Crossref] [PubMed]
- Comber JD, Philip R. MHC class I antigen presentation and implications for developing a new generation of therapeutic vaccines. Ther Adv Vaccines 2014;2:77-89. [Crossref] [PubMed]
- Caron E, Kowalewski DJ, Chiek Koh C, et al. Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Mol Cell Proteomics 2015;14:3105-17. [Crossref] [PubMed]
- Sadelain M, Brentjens R, Rivière I. The promise and potential pitfalls of chimeric antigen receptors. Curr Opin Immunol 2009;21:215-23. [Crossref] [PubMed]


