|
Predicting survival and recurrence by gene expression
profiling
GEP has been used to predict response to treatment
and patients’ outcome ( 13, 31, 44-68). Beer et al. analyzed
the genetic profile in 86 patients with primary lung
adenocarcinoma, and found that the genes most associated
with survival were identified to create a risk index based
on the top 50 genes that separated patients into high-risk
and low-risk groups. When applying this risk predictor to a
test data set of 62 stage I patients from another study, they
were able to predict survival with statistical significance
difference (P=0.006) ( 51). This study also identified certain
patients with stage I along with stage III disease with poor
prognosis based on gene profile. This demonstrated the
ability for GEP to identify a patient with poor prognosis
that is independent of the stage at the time of diagnosis. Guo et al. devised a computational model system
that redicted the clinical outcome of individual patients
based on their GEP. A 37-gene signature was created,
and the authors studied a cohort of 86 patients diagnosed
with lung adenocarcinoma. The gene signature was then
applied to predict the survival of the other 84 patients
with adenocarcinoma. The predictive accuracy of the gene
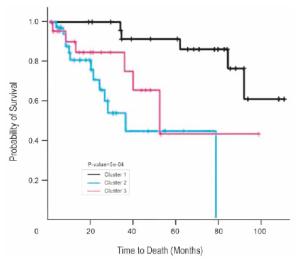
signature was 96%. The cluster analysis, using the 37-gene
signature, aggregated the test patient samples into 3 groups
with good (mean survival, 66.9 months), moderate (mean
survival, 27.6 months), and poor (mean survival, 22.4
months) prognoses (Kaplan-Meier analysis; P < .0005; logrank
test) ( Fig 1). Notably, when the results were reviewed,
all patients who had grouped together in cluster 1 (good
prognosis) had stage I disease, with N0 lymph node status
(no metastasis) and smaller tumor size (T1 or T2) ( 63). Landmark studies such as the one conducted by Potti
et al. from Duke University have identified GEP, which
predicted the risk of recurrence following surgery from
a cohort of patients with early-stage NSCLC ( 52). The
accuracy was > 70%. The investigators were also able to
identify a subgroup of patients with stage IA disease who
were at high risk for recurrence, with a very poor survival,
and who might be suitable for adjuvant chemotherapy. This is clinically relevant when the current standard of care for
patients with stage IA disease is just clinical observation
(no adjuvant chemotherapy is offered) because of a 70%
chance of 5-year survival. This genetic strategy was
then validated in two separate cohorts from multicenter
cooperative group trials: 25 patients from the American
College of Surgeons Oncology Group Z0030 study and
84 from the prospective CALGB 9761 trial, this genomic
strategy had an overall predictive accuracy of 72 and
79%, respectively. This gene expression profile also was
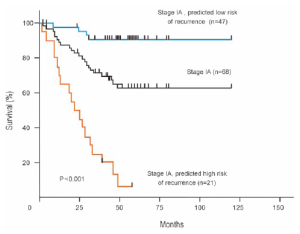
applied to 68 patients with stage IA disease, who are not
usually candidates for adjuvant chemotherapy. Kaplan-
Meier survival curves were generated for the group as a
whole and for the subgroups predicted to be at high or low
risk for recurrence by the lung metagene model. Although
the survival rate for the group was approximately 70% at
4 years, the survival rate for those predicted to be at low
risk was 90% and less than 10% for those predicted to be
at high risk, thus identifying the subgroup of patients with
stage IA NSCLC at high risk of recurrence, who might
benefit from adjuvant chemotherapy ( Fig 2). In another important study from Taiwan University
( 13), authors examined the expression of multiple genes
associated with invasive activity in frozen specimens of
lung-cancer tissue from 125 randomly selected patients
who underwent surgical resection of NSCLC and not
received adjuvant chemotherapy, to identify a gene
signature that is correlated with clinical outcome. Sixteen genes were initially identified by analyzing
microarray data and then confirmed by RT-PCR. From
these, the authors further identified five genes that were
significantly associated with survival. The levels of
expression of these five genes were used to construct a
decision tree to classify patients as having a high-risk gene
signature or a low-risk gene signature. The five selected
genes were: dual-specificity phosphatase 6 (DUSP6),
monocyte-to-macrophage differentiation-associated protein
(MMD), signal transducer and activator of transcription
1 (STAT1), v-erb b2 avian erythroblastic leukemia viral
oncogene homolog 3 (ERBB3), and lymphocyte-specific
protein tyrosine kinase (LCK).
The authors identified 59 patients with high-risk
gene signatures and 42 with low-risk gene signatures,
according to gene expression as measured with RT-PCR
and decision-tree analysis. The five-gene signature was
strongly associated with OS (sensitivity 98%; specificity
93%; positive predictive value 95%; negative predictive
value 98%; and overall accuracy 96%). The presence of
a high-risk five-gene signature in the NSCLC tumors
was associated with an increased risk of recurrence and
decreased OS. With a median follow-up of 20 months,
the patients with a high-risk gene signature had a shorter
median OS than the patients with a low-risk gene signature
(20 months versus 40 months, P<0.001). The high-risk gene
signature was associated with a median RFS of 13 months,
whereas the low-risk gene signature was associated with a median RFS of 29 months (P=0.002).
According to multivariate regression analysis, the
high-risk five-gene signature, tumor stage III and older
age were significantly associated with death from any
cause among the 101 patients, and the high-risk five-gene
signature and tumor stage III were significantly associated
with recurrence of cancer as well (HR for the high-risk
signature versus the low-risk signature, 1.92; 95% CI, 1.06
–3.46; P=0.03). In a subgroup analysis of 59 patients with
stage I or II disease, those with a high-risk gene signature
had a shorter OS and a shorter RFS than those with a lowrisk
gene signature. These results were validated in an
independent cohort of 60 patients with NSCLC and with
the use of a set of published microarray data from 86
patients from a Western population with NSCLC.
The identif ication of f ive genes that are closely
associated with the outcomes in patients with NSCLC
has clinical implications. Patients who have tumors
with a high-risk gene signature could benefit from a
cisplatin-based adjuvant chemotherapy, whereas those
with a low-risk gene signature could be spared what
may be unnecessary treatment. Prospective, large scale,
multicenter studies are necessary to test this idea. These
five genes that can predict the clinical outcome in patients
with NSCLC may also reveal targets for the development
of therapy for lung cancer. STAT1 causes arrested growth
and apoptosis in many types of cancer cells by inducing
the expression of p21WAF1 and caspase ( 53, 54). MMD is
preferentially expressed in mature macrophages ( 55). Some
studies have shown that macrophage activation promotes
cancer metastasis ( 56), although the function of the MMD
protein is unknown. DUSP6 inactivates extracellular
signal-regulated kinase 2 (also known as mitogen-activated
protein kinase 1), resulting in tumor suppression and
apoptosis ( 57). ERBB3, a member of the epidermal growth
factor receptor family of tyrosine kinases, can shorten cell
survival ( 58). LCK, a member of the Src family of protein
tyrosine kinases, is expressed mainly in T cells and is one
of the first signaling molecules downstream of the T-cell
receptor. It plays a key role not only in the differentiation
and activation of T cells but also in the induction of
apoptosis ( 59). In addition, LCK is expressed in many
cancers and regulates the mobility of cancer cells ( 60, 61). Bianchi et al. proposed a qRT-PCR–based 10-gene
molecular signature for adenocarcinoma ( 46). They selected
49 unbiased genes based on a meta-analysis of previously
published adenocarcinoma microarray data and combined
this with a biased set of 31 additional genes selected from
the literature demonstrated to either be important for
tumorigenesis and/or to represent prognostic lung cancer
markers. These 80 genes were tested on a training cohort
of stage I adenocarcinoma patients using a leaveone-out validation model yielding a 10-gene signature. In two
separate validation cohorts of stage I adenocarcinoma
patients, this 10-gene signature was more accurate than
stage (IA vs. IB), age, sex, differentiation, or presence
of a K-ras mutation in predicting survival. In addition, it
also demonstrated differences in survival when applied
to separate cohorts of stage IA and stage IB patients with
adenocarcinoma but, similar to the findings by Chen et al.
( 13), did not demonstrate significant predictive differences
in stage II or III adenocarcinomas. Lau et al. proposed a qRT-PCR–based 3-gene signature
for NSCLC ( 45). One hundred twenty-eight candidate genes
were identified using data from 7 previous microarray
based profiling studies and assayed by qRT-PCR in 147
frozen NSCLC samples. Using a statistical method based
on concordance index and risk scores, a 3-gene signature
(STX1A, CCR7, and HIF1A) was developed. When
applied to their own training cohort as well as to two
cohorts from previously published microarray data sets,
they demonstrated a statistically significant difference in
survival between patients with stage I NSCLC classified
as having either good or poor prognosis. In agreement
with the above studies, this difference did not hold true for
patients with stage II disease. They also demonstrated that
their 3-gene signature was better at predicting survival in
their training cohort stage I patients than stage, histology,
or smoking status. Skrzypski et al. examined the expression pattern of
29 genes selected by cDNA studies to test their clinical
prognostic value in early-stage squamous cell carcinoma
(SCC) of the lung ( 49). From 2000 to 2004, freshly frozen
primary tumor specimens were obtained at the time of
the surgery from 66 SCC patients and gene expression of
the 29 genes was assessed by quantitative RT-PCR using
low-density arrays. Expression values were dichotomized
using the median value as the cutoff. The univariate
analysis showed 10 genes with prognosis value: PH4
(P=0.01); macrophage-colony stimulating factor (CSF1),
which attracts macrophages and induce them to express
EGF (P= 0.002); EGFR (P=0.05); KIAA0974 (P=0.02);
ANLN (P=0.02); carbonic anhydrase IX (CA IX), which is
regulated by hypoxia and plays a role in chemoresistance
(P=0.007); VEGFC (P=0.03); neurotrophic tyrosine
receptor kinase 1 (P=0.04); fibronectin (P=0.002); insulin
receptor (P= 0.03). In the multivariate analysis of survival,
CSF1, EGFR and CA IX, and tumor size emerged as
significant variables (P=0.005, 0.02, <0.0001, 0.02,
respectively). Roepman et al. aimed to develop a gene expression
prof ile for stage I and stage II NSCLC, allowing
identification of patients with a high risk of disease
recurrence within 2 to 3 years after initial diagnosis. Whole-genome gene expression microarrays were used to
analyze frozen tumor samples from 172 NSCLC patients
(pT1-2, N0-1, M0) from five European institutions, who
had undergone complete surgical resection. A 72-gene
expression prognostic NSCLC classifier was developed.
Based on the classifier score, patients were classified as
either high or low risk of disease recurrence. Patients
classified as low risk showed a significantly better
recurrence-free survival both in the training set (P < 0.001;
n = 103) and in the independent validation set (P < 0.05; n
= 69). It was found that the 72-gene signature was closely
associated with recurrence-free and overall survival in
early-stage NSCLC patients and may become a tool for
patient selection for adjuvant therapy ( 62). Reed et al. proposed a qRT-PCR–based 2-gene signature
for adenocarcinoma ( 48). Pooling microarray analysis of
NSCLC cell lines in conjunction with correlation mapping
of genes highly expressed in other tumors produced 14
candidate genes. These genes were tested by qRT-PCR on
20 adenocarcinoma samples yielding a 2-gene signature
(CK19 and EpCAM2). This 2-gene signature revealed
survival differences in high- and low-risk patients in their
training cohort (HR, 6.2) and in a separate validation
cohort (HR, 4.5) by Kaplan-Meier analysis. Raz et
al. proposed a qRT-PCR–based 4-gene signature for
adenocarcinoma ( 50). Seventy-six cancer-related candidate
genes were selected from 217 genes demonstrated to have
prognostic significance in previously published studies
by content experts and literature review. Sixty-one of
these genes for which reliable qRT-PCR data could be
produced were assayed using qRT-PCR in a cohort of
120 adenocarcinoma samples. Cross-validation using
Cox proportional hazards regression supported a 4-gene
signature (WNT3A, RND3, LCK, and ERBB3). When
applied to a cross-validated cohort of 70 patients with stage
I adenocarcinoma, statistically significant differences in
OS (87% vs. 38%) and disease-free survival (77% vs. 35%)
were shown for high- and low-risk patients. This compared
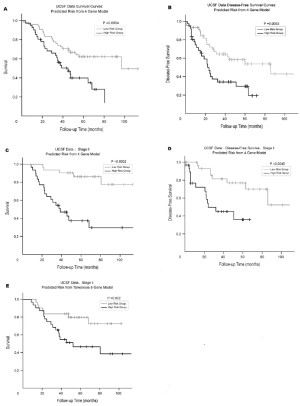
favorably with the 5-gene signature of Chen et al. ( 13) ( Fig
3). When applied to the Raz et al cross-validated cohort,
the signature developed by Chen et al demonstrated 5-year
OS of 80% and 47%, respectively, for high- and lowrisk
patients. Notably, 2 of the genes (ERBB3 and LCK)
overlapped between the Raz et al and Chen et al signatures. Identified prognostic classifiers for early-stage NSCLC
indicated large differences in sample numbers, microarray
platform, and classifier design. Although a great variety
of statistical models have been used, the performance of
the different classifiers is similar with overall accuracies
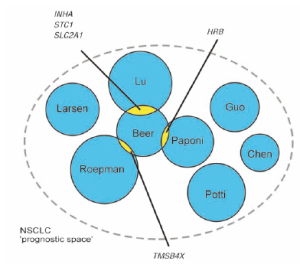
between 70% and 80% and a hazard ratio of 3 to 4. The
overlap in profile genes, however, is limited to only 5
of a total of 327 genes ( Fig. 4) even though it includes two studies ( 63, 64) that reanalyzed existing data ( 51)
but showed respectively no and three genes in overlap
( 62). Ein-Dor and coworkers ( 65) demonstrated that
biological heterogeneity leads to thousands of samples
being required to identify robust and reproducible
subsets for most tumor types. These conclusions are
supported by the finding that thousands of genes display
intratumor heterogeneity, likely caused by the diversity
of tumor microenvironments and cell populations ( 66, 67).
However, Boutros and coworkers hypothesized that
differing statistical methodologies contribute to this lack
of overlap ( 68). To test the hypothesis, they analyzed our
previously published quantitative RT-PCR dataset with a
semisupervised method. A 6-gene signature was identified
and validated in 4 independent public microarray datasets
that represent a range of tumor histologies and stages. The
result demonstrated that at least 2 prognostic signatures
can be derived from this single dataset. They further
estimated the total number of prognostic signatures in this
dataset with a 10-millionsignature permutation study. Their
6-gene signature was among the top 0.02% of signatures
with maximum verifiability, reaffirming its efficacy.
Importantly, the analysis identified 1,789 unique signatures,
implying that their dataset contains >500,000 verifiable
prognostic signatures for NSCLC. The result appears to
rationalize the observed lack of overlap among reported
NSCLC prognostic signatures ( 68).
|
|
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin 2007;57:43-66.[LinkOut]
- Ponn RB, Lo Cicero J III, Daly BDT. Surgical treatment of non small cell lung cancer. In: Shields TW, Lo Cicero J III, Ponn R, Rusch VW, Editors. General Thoracic Surgery, 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2005. p. 1548-87.
- Wright G, Manser RL, Byrnes G, Hart D, Campbell DA. Surgery for early-stage non-small cell lung cancer: systematic review and metaanalysis of randomised controlled trials. Thorax 2006;61:597-603.[LinkOut]
- Strauss GM. Adjuvant chemotherapy of lung cancer: methodologic issues and therapeutic advances. Hematol Oncol Clin North Am 2005;19:263-81.[LinkOut]
- Nesbitt JC, Putnam JB Jr, Walsh GL, Roth JA, Mountain CF. Survival in early stage non-small cell lung cancer. Ann Thorac Surg 1995;60:466-72.[LinkOut]
- Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet 2000;355:479-85. [Erratum, Lancet 2000;355:1280.]
- Hoang T, Xu R, Schiller JH, Bonomi P, Johnson DH. Clinical model to predict survival in chemonaïve patients with advanced nonsmall-cell lung cancer treated with third-generation chemotherapy regimens based on eastern cooperative oncology group data. J Clin Oncol 2005;23:175-83.[LinkOut]
- Forrest LM, McMillan DC, McArdle CS, Angerson WT, Dagg K, Scott HR. A prospective longitudinal study of performance status, an inflammation-based score (GPS) and survival in patients with inoperable non-small-cell lung cancer. Br J Cancer 2005;92:1834-6.[LinkOut]
- Brundage MD, Davies D, Mackillop WJ. Prognostic factors in non-small cell lung cancer: a decade of progress. Chest 2002;122:1037-57.[LinkOut]
- Meyerson M, Carbone DP. Genomic and proteomic profiling of lung cancers: lung cancer classification in the age of targeted therapy. J Clin Oncol 2005;23:3219-26.[LinkOut]
- Custodio AB, González-Larriba JL, Bobokova J, Calles A, Alvarez R, Cuadrado E, et al. Prognostic and predictive markers of benefit from adjuvant chemotherapy in early-stage non-small cell lung cancer. J Thorac Oncol 2009;4:891-910.[LinkOut]
- Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 2005;5:845-56.[LinkOut]
- Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 2007;356:11-20.[LinkOut]
- Endoh H, Tomida S, Yatabe Y, Konishi H, Osada H, Tajima K, et al. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol 2004;22:811-9.[LinkOut]
- Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, et al. A genomic strategy to refine prognosis in early-stage nonsmall-cell lung cancer. N Engl J Med 2006;355:570-80.[LinkOut]
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420:860-7.[LinkOut]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683-92.[LinkOut]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature 2008;454:436-44.[LinkOut]
- Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature 2008; 452:553-63.[LinkOut]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995;270:467-70.[LinkOut]
- Lashkari DA, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, et al. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci U S A 1997; 94:13057-62.[LinkOut]
- Ramaswamy S. Translating cancer genomics into clinical oncology. N Engl J Med 2004; 350:1814-6.[LinkOut]
- Paik S, Kim CY, Song YK, Kim WS. Technology insight: Application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat Clin Pract Oncol 2005;2:246-54.[LinkOut]
- Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J Clin Epidemiol 2007; 60:1205-19.[LinkOut]
- Kratz JR, Jablons DM. Genomic prognostic models in early-stage lung cancer. Clin Lung Cancer 2009;10:151-7.[LinkOut]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105-11.[LinkOut]
- Heguy A, O'Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey B-G, et al. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J Mol Med 2006;84:318.[LinkOut]
- Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol 2003;163:1949-60.[LinkOut]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503-21.[LinkOut]
- Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, et al. Vinorelbine plus Cisplatin vs. Observation in resected nonsmall-cell lung cancer. N Engl J Med 2005;352:2589-97.[LinkOut]
- Tsao MS, Zhu C, Ding K, Strumpf D, Pintilie M, Meyerson M, et al. A 15-gene expression signature prognostic for survival and predictive for adjuvant chemotherapy benefit in JBR. 10 patients. J Clin Oncol 2008;26(supp):abstr 7510.
- Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A 2001;98:10787-92.[LinkOut]
- Grever MR, Schepartz SA, Chabner BA. The National Cancer Institute: cancer drug discovery and development program. Semin Oncol 1992; 19:622-38.[LinkOut]
- Stinson SF, Alley MC, Kopp WC, Fiebig HH, Mullendore LA, Pittman AF, et al. Morphological and immunocytochemical characteristics of human tumor cell lines for use in a disease-oriented anticancer drug screen. Anticancer Res 1992;12:1035-53.[LinkOut]
- Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med 2006;12:1294-300. Epub 2006 Oct 22.[LinkOut]
- Beane J, Spira A, Lenburg ME. Clinical impact of high-throughput gene expression studies in lung cancer. J Thorac Oncol 2009;4:109-18.[LinkOut]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500.[LinkOut]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005; 353:123-32.[LinkOut]
- Rosell R, Taron M, Sanchez JJ, Paz-Ares L. Setting the benchmark for tailoring treatment with EGFR tyrosine kinase inhibitors. Future Oncol 2007; 3:277-83.[LinkOut]
- O'Byrne KJ, Danson S, Dunlop D, Botwood N, Taguchi F, Carbone D, et al. Combination therapy with gefitinib and rofecoxib in patients with platinum-pretreated relapsed non small-cell lung cancer. J Clin Oncol 2007; 25:3266-73.[LinkOut]
- Okano T, Kondo T, Fujii K, Nishimura T, Takano T, Ohe Y, et al. Proteomic signature corresponding to the response to gefitinib (Iressa, ZD1839), an epidermal growth factor receptor tyrosine kinase inhibitor in lung adenocarcinoma. Clin Cancer Res 2007;13:799-805.[LinkOut]
- Altorki N, Lane ME, Bauer T, Lee PC, Guarino MJ, Pass H, et al. Phase II proof-of-concept study of pazopanib monotherapy in treatment-naive patients with stage I/II resectable non-small-cell lung cancer. J Clin Oncol 2010;28:3131-7. Epub 2010 Jun 1.[LinkOut]
- Nikolinakos PG, Altorki N, Yankelevitz D, Tran HT, Yan S, Rajagopalan D, et al. Plasma cytokine and angiogenic factor profiling identifies markers associated with tumor shrinkage in early-stage non-small cell lung cancer patients treated with pazopanib. Cancer Res 2010;70:2171-9. Epub 2010 Mar 9.[LinkOut]
- Tomida S, Koshikawa K, Yatabe Y, Harano T, Ogura N, Mitsudomi T, et al. Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene 2004;23:5360-70.[LinkOut]
- Lau SK, Boutros PC, Pintilie M, Blackhall FH, Zhu CQ, Strumpf D, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol 2007;25:5562-9.[LinkOut]
- Bianchi F, Nuciforo P, Vecchi M, Bernard L, Tizzoni L, Marchetti A, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest 2007;117:3436-44.[LinkOut]
- Seike M, Yanaihara N, Bowman ED, Zanetti KA, Budhu A, Kumamoto K, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst 2007;99:1257-69.[LinkOut]
- Reed CE, Graham A, Hoda RS, Khoor A, Garrett-Mayer E, Wallace MB, et al. A simple two-gene prognostic model for adenocarcinoma of the lung. J Thorac Cardiovasc Surg 2008;135:627-34.[LinkOut]
- Skrzypski M, Jassem E, Taron M, Sanchez JJ, Mendez P, Rzyman W, et al. Three-gene expression signature predicts survival in early-stage squamous cell carcinoma of the lung. Clin Cancer Res 2008;14:4794-9.[LinkOut]
- Raz DJ, Ray MR, Kim JY, He B, Taron M, Skrzypski M, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res 2008;14:5565-70.[LinkOut]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816-24. Epub 2002 Jul 15.[LinkOut]
- Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, et al. A genomic strategy to refine prognosis in early-stage nonsmall-cell lung cancer. N Engl J Med 2006;355:570-80.[LinkOut]
- Yu H, Jove R. The STATs of cancer-new molecular targets come on age. Nat Rev Cancer 2004;4:97-105.[LinkOut]
- Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science 1997;278:1630 -2.[LinkOut]
- Rehli M, Krause SW, Schwarzfischer L, Kreutz M, Andreesen R. Molecular cloning of a novel macrophage maturation-associated transcript encoding a protein with several potential transmembrane domains. Biochem Biophys Res Commun 1995;217:661- 7.[LinkOut]
- Chen JJ, Lin YC, Yao PL, Yuan A, Chen HY, Shun CT, et al. Tumor-associated macrophages: the double-edged sword in cancer progression. J Clin Oncol 2005;23:953-64.[LinkOut]
- Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol 2003;162:1807-15.[LinkOut]
- Müller-Tidow C, Diederichs S, Bulk E, Pohle T, Steffen B, Schwäble J, et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res 2005;65:1778-82.[LinkOut]
- Zamoyska R, Basson A, Filby A, Legname G, Lovatt M, Seddon B. The influence of the src-family kinases, on T cell differentiation, survival and activation. Immunol Rev 2003;191:107-18.[LinkOut]
- Mahabeleshwar GH, Kundu GC. Tyrosine Kinase p56lck regulates cell motility and nuclear factor kappa B-mediated secretion of urokinase type plasminogen activator through tyrosine phosphorylation of IkappaBalpha following hipoxia/reoxygenation. J Biol Chem 2003;278:52598-612.[LinkOut]
- Mahabeleshwar GH, Das R, Kundu GC. Tyrosine Kinase, p56lckinduced cell motility, and urokinase-type plasminogen activator secretion involve activation of epidermal growth factor receptor/extracellular regulated kinase pathways. J Biol Chem 2004;279:9733-42.[LinkOut]
- Roepman P, Jassem J, Smit EF, Muley T, Niklinski J, van de Velde T, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res 2009;15:284-90.[LinkOut]
- Guo L, Ma Y, Ward R, Castranova V, Shi X, Qian Y. Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clin Cancer Res 2006;12:3344-54.[LinkOut]
- Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med 2006;3:e467.[LinkOut]
- Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA 2006;103:5923-8.[LinkOut]
- Bachtiary B, Boutros PC, Pintilie M, Shi W, Bastianutto C, Li JH, et al. Gene expression profiling in cervical cancer: An exploration of intratumor heterogeneity. Clin Cancer Res 2006;12:5632-40.[LinkOut]
- Blackhall FH, Pintilie M, Wigle DA, Jurisica I, Liu N, Radulovich N, et al. Stability and heterogeneity of expression profiles in lung cancer specimens harvested following surgical resection. Neoplasia 2004;6:761-7.[LinkOut]
- Boutros PC, Lau SK, Pintilie M, Liu N, Shepherd FA, Der SD, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A 2009;106:2824-8.[LinkOut]
Cite this article as: Shao WL, Wang DY, He JX. The role of gene expression profiling in early-stage non-small cell lung cancer. J Thorac Dis 2010;2(2):89-99. doi: 10.3978/j.issn.2072-1439.2010.02.02.010
|