Insight into the regulatory mechanism of VEGFA isoforms in p53 mutant breast cancer: can it be substantiated with solely in vitro studies?
Breast cancer is now the leading type of cancer in women, and it’s the primary form of cancer-related death. Five different subtypes of breast cancer have been identified according to some molecular profiling studies. They are normal breast-like, basal-like, luminal A, luminal B, and ERBB2+ breast cancer respectively (1). The investigation and application of neoadjuvant and potential treatment targets in breast cancer have been popular in recent years, as well as the signaling pathways and molecular mechanism exploration in the development of breast cancer. Based on the discovery of potential molecular targets, it is increasingly important to identify the relationship between targets and clinical diseases, and to evaluate and begin clinical trials for the targeted therapy subsequently, also known as “translational medicine”. Currently, the regulatory mechanisms for some genes and proteins have been well defined in breast cancer and other types of cancer research. p53 (TP53) is one of the most studied genes involved in cancer formation and progression. As a tumor suppressor gene, p53 participates in cell cycle arrest, apoptosis, senescence and DNA repair (2). The p53 gene has been found to be the most frequently mutated gene in triple-negative breast cancer, driving cancer formation and progression (2,3). Therefore, mutant p53 becomes a high-priority target for anticancer therapy. The long non-coding RNAs (lncRNAs), which are characterized by high specificity and are easily detected in tumor tissues, can regulate gene transcription and basal transcription machinery, and participate in post-transcriptional as well as epigenetic regulation. Altered expression of lncRNAs has been implicated in many cancers. LncRNA has shown potential as a biomarker in the diagnosis and prognosis of cancers including breast cancer (4). Moreover, splicing factors have attracted increasing attention in cancer research, since they play a critical role in regulating alternative splicing, which is further involved in apoptotic regulation and programmed cell death in cancer (5).
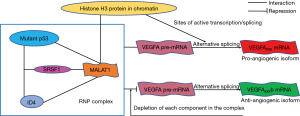
Pruszko and colleagues report one kind of vascular endothelial growth factor A (VEGFA) regulatory mechanism mediated by lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) in breast cancer cells expressing gain-of-function mutant p53 and ID4 proteins (6). The authors identified “a quaternary ribonucleoprotein (RNP) complex comprising the MALAT1 lncRNA and the SR splicing factor 1 (SRSF1) oncogenic splicing factor, as well as mutant p53 and ID4 proteins”. This RNP complex could control VEGFA isoforms and sustain angiogenic potential in breast cancer cells by influencing the alternative splicing of VEGFA pre-mRNA and further repressing the expression of anti-angiogenic VEGFA isoforms. They emphasized SRSF1 as a required factor for the interaction between MALAT1 and the mutant p53-ID4. Further, “a reduction in the angiogenic potential of breast cancer cells” had been shown when “the depletion of MALAT1 or of any of the protein components of this RNP complex” performed. Finally, the authors concluded that “The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1”. Thus, the disassembling of the RNP complex comprising mutant p53/ID4/SRSF1/MALAT1 might hold therapeutic premise for mutant p53 breast cancers.
MALAT1 is among the most abundant lncRNAs, and altered MALAT1 expression has been confirmed to be involved in numerous cancers (7). MALAT1 regulates alternative splicing of endogenous pre-mRNAs by interacting with SRSF1, SRSF2 and SRSF3 directly and modulating SRSF phosphorylation (8,9). Meanwhile, p53 has been shown to be a major downstream mediator of MALAT1 activity (10), and can be stabilized by SRSF1 through ribosomal protein L5 (RPL5) (11). The interactions among lncRNA MALAT1 and WT/mutant p53, ID4, SRSF1 are fairly complicated. In this paper, a number of techniques were employed to verify involvement of each component of the targeted complex, including the proximity ligation assay (PLA), RNA FISH and several co-immunoprecipitations followed by either immunoblot or qRT-PCR analysis. The authors revealed a novel insight into the complex interactions mentioned above, and delineated their roles in the regulation of the downstream target VEGFA isoforms (Figure 1).
There are some limitations to this study. First, there was no animal work for this study, unfortunately. The authors argued that the RNP complex comprising mutant p53/ID4/SRSF1/MALAT1 could favor angiogenesis activity in breast cancer with p53 mutation. However, it is well established that the tumor microenvironment and tumor cell status in vivo differ from in vitro. Different conclusions might be drawn as a result of in vivo animal trials. Testing a series of mice xenograft breast cancer models could make the conclusion more complete and reasonable. In addition, extra data could be achieved simultaneously, including the observation of angiogenesis activity in tumor tissue, the evaluation of tumor biological behavior with intact or impaired RNP complex, and so on in mice xenograft models. The completion of in vivo experiments section might profoundly improve the value of this study. Second, this study did not contribute significantly to translational medicine. The primary goal for medical research is to make progress to clinical trials. The authors did not perform any clinical research of their own, and instead collected gene expression data from the METABRIC cohort and online MSigDB database to illustrate the association of VEGFA signature expression with ID4 expression in mutant p53-carrying basal-like breast cancers. Theoretically, if the authors collected clinical follow-up data and pathological tissues of breast cancer patients, they might resolve the following issues: clarifying the location and quantity of the target genes expression; and confirming the difference of expression level of the key genes not only between breast cancer tissue and non-cancerous breast tissue, but also identifying the association between target genes expression and clinicopathological features such as regional lymph node metastasis and prognostic implications in the different breast cancer subtypes. Solving these issues is the vital part in translational medicine, which could contribute much better to the research of clinical treatment to breast cancer.
In summary, although the authors are to be commended for displaying a novel regulatory mechanism to VEGFA isoforms in p53 mutant breast cancer cells mediated by a RNP complex comprising mutant p53/ID4/SRSF1/MALAT1, additional studies in vivo and related in translational medicine are critical to obtaining a more persuasive and substantial conclusion.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003;100:8418-23. [Crossref] [PubMed]
- Duffy MJ, Synnott NC, Crown J. Mutant p53 as a target for cancer treatment. Eur J Cancer 2017;83:258-65. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer 2017;140:1955-67. [Crossref] [PubMed]
- Kędzierska H, Piekiełko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett 2017;396:53-65. [Crossref] [PubMed]
- Pruszko M, Milano E, Forcato M, et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep 2017;18:1331-51. [Crossref] [PubMed]
- Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 2016;30:34-51. [Crossref] [PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. [Crossref] [PubMed]
- Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012;18:1487-99. [Crossref] [PubMed]
- Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 2013;9:e1003368. [Crossref] [PubMed]
- Fregoso OI, Das S, Akerman M, et al. Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol Cell 2013;50:56-66. [Crossref] [PubMed]


