Long non-coding RNA DANCR promotes cell proliferation, migration, invasion and resistance to apoptosis in esophageal cancer
Introduction
Esophageal cancer is one of the most prevalent malignant tumors with high morbidity and mortality, and it ranks as the sixth-leading cause of tumor-relevant death in the world (1-3). As a general common sense, esophageal cancer is mainly divided into esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (4,5). China has one of the world’s highest incidence rates of this disease due to various factors, including diet, environment and modern life-styles (6,7). It is hard to achieve satisfactory therapeutic effect by using traditional treatments such as chemotherapy, radiation therapy, surgical treatment, and comprehensive treatment for esophageal cancer (8-10). In addition, the reported recurrence rate of this carcinoma was larger than 40%, and its 5-year survival rate was 20–30% for patients received traditional treatments (11,12). The pathology of esophageal cancer is complicated that involves a series of physiological changes including abnormal proliferation, metastasis, invasion and apoptosis of cells (13,14). It is universality accepted that the major cause of cancer-related death is complications resulting from tumor metastasis (15,16). Hence, there is an urgent need for novel therapeutic strategies for esophageal cancer treatment (17).
Long non-coding RNAs (lncRNAs) are transcribed molecules with a length over 200 nucleotides, but are not protein-coding (18,19). LncRNAs, had been proved to involved in multiple life proceed, such as cell proliferation, migration and apoptosis (18,20,21). Based on the importance of lncRNAs in biological processes, we investigated the lncRNA DANCR. It was reported that DANCR could inhibited the epidermal cells’ differentiation and played an important role in maintaining stem cell properties of hepatocellular carcinoma (HCC) cells (22,23). However, the biological roles and influences of DANCR in esophageal cancer have not been indicated yet. In this study, we detected the expression of DANCR in ESCC tissues and ESCC ECA109 cell. We also analyzed the regulatory mechanisms of DANCR on proliferation, migration, invasion, apoptosis and cell cycle of ECA109 cells in vitro. Our results indicated that DANCR could have potentially beneficial effects on preventing the metastasis of ESCC, thus providing a necessary experimental basis for further clinical research.
Methods
Patients and tissue specimens
In the study, total 32 patients with ESCC were recruited in the Affiliated Hospital of Nantong University from 2015 to 2016. All diagnoses were based on histopathological evidence. All patients were not received preoperative treatments for cancer, such as radiotherapy, chemotherapy or immunotherapy. All fresh tissues (ESCC tissues and adjacent tissues) after surgical separation were immediately washed with sterile phosphate-buffered saline (PBS) before stored at −80 °C until total RNA was extracted. This study was approved ethically by the Committee of Affiliated Hospital of Nantong University (No. 2015-070). All patients were notified consent for the use of their tissues for scientific research.
Cell culture and cell transfection
The human ESCC cell lines ECA109 and TE-1 were supplied by the cell resource center of Shanghai institute for biological sciences, Chinese Academy of Sciences. Cell culture was performed in DMEM high glucose Medium (ThermoFisher Scientific, Waltham, MA, USA) added with 10% fetal calf serum (FCS) (GIBCO, Carlsbad, CA, USA) and 100 U/mL penicillin-streptomycin mixture (Beyotime Institute of Biotechnology, Haimen, China) in CO2 incubator 5% CO2 at 37 °C. SiRNA targeting DANCR (siDANCR) was synthesized by RiboBio company (Guangzhou, China). The applied siRNA sequences for DANCR were as follows: 5'-UCGGAGGUGGAUUCUGUUA-3' (sense) and 5'-UUUCUGUUUCCUCCUCCGU-3' (antisense). Cells were transfected with 100 nM negative control (NC) siRNA (scramble) (siDANCR-NC) or siRNA targeting DANCR using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
RNA extraction and quantitative real-time PCR (qRT-PCR)
The Trizol reagent (ThermoFisher Scientific, Carlsbad, CA) was used to extract the total RNA from ESCC tissues, adjacent normal tissues, ECA109 cells and TE-1 cells in accordance with the operation manual. Then, total RNA was reverse transcripted using a Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA). Based on the operation manual, qRT-PCR with the SYBR Green Master Mixture (Roche, Basel, Switzerland) was used to analyze the expression of DANCR in ESCC tissues and ESCC cells in triplicate. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a quantitative normalization. The sequences of primers were described as below: 5'-GCGCCACTATGTAGCGGGTT-3' (upstream for DANCR), 5'-TCAATGGCTTGTGCCTGTAGTT-3' (downstream for DANCR), 5'-AGAAGGCTGGGGCTCATTTG-3' (upstream for GAPDH) and 5'-AGGGGCCATCCACAGTCTTC-3' (downstream for GAPDH). Each sample was evaluated in triplicate and then used for the analysis of the relative transcription data using the 2−ΔΔCT method.
MTT assay
ECA109 cells were seeded into 96-well plates in triplicate at a density of 5×103 cells per well. Cell viability was tested using MTT assay (Beyotime Institute of Biotechnology, Haimen, China) 48 h after transfected with 100 nmol siDANCR or 100 nmol siDANCR-NC. Three replicates for each group. Each well was added with 10 µL MTT solution (5 mg/mL) and 100 µL DMEM medium with 10% FCS, continuously further incubation for 4 h at 37 °C. The culture medium was then replaced with 100 µL of DMSO (Sigma, St. Louis, MO, USA). The optical density (OD) was measured at 570 nm by ELX-800 microplate reader (BioRad, Hercules, CA, USA) after crystal was fully dissolved.
Wound healing assay
Cells were seeded into 24-well plates after transected. Scratch wounds were manipulated with the tip of a 100 µL pipette to scrub the cell using when the cell density reached above 90%. Then the cells were washed twice with 0.01% PBS and fresh serum-medium was added to further incubation at 37 °C and 5% CO2. Photos were taken randomly at 0, 24 and 48 h using microscopy to evaluate the ability of cell migration. Independent experiments were repeated three times.
Cell migration and invasion assay
For the migration assay, cells were collected and resuspended in DMEM and then 200 µL suspension (105 cells) was placed in Transwell chambers (CORNING, Corning, NY, USA) in 24-well plates 48 h after transfected. The lower chamber was added with 500 µL DMEM media supplemented with 10% FCS. After culture for 24 h, the cells in the upper surface were wiped out using cotton swabs. However, the cells in the lower surface were fixed with 4% paraformaldehyde (PFA), and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology, Haimen, China) for 40 min. The cells in the lower surface were just migrated cells. During the invasion test, firstly 50 µL Matrigel matrix was added into the upper surface before adding the cells and other conditions were the same as the migration assay described above. We counted five randomly selected fields to calculate the average cell number in triplicate.
TUNEL assay
ECA109 cells were seeded into 24-well plates with glass coverslips with 5×104 in each well 48 h after transfected. Cells were fixed with 4% PFA fixation solution for 20 min at 15–25 °C after incubation for 24 h, and washed 30 min with 0.01% PBS. After that, cells were incubated with blocking solution for 10 min at 15–25 °C. Coverslips were rinsed with 0.01% PBS twice and incubated in permeabilization solution for 2 min on ice. The total volume (5 µL) of enzyme solution was added to the remaining 45 µL label solution to obtain 50 µL TUNEL (Roche, Basel, Switzerland) mixture for each coverslip, and mix well to equilibrate components. Coverslips were allowed to incubate with it for 1 h at 37 °C. Finally, they were incubated with Hoechst 33258 (Sigma-Aldrich, St. Louis, MO) for 10 min followed by examination under a fluorescence microscope (Zeiss, Oberkochen, Germany).
Flow cytometry (FCM) assay
Cells were collected after transected and washed with 0.01% PBS twice. Afterwards, 195 µL binding buffer was added to resuspend the cells, and 5 µL of Annexin V-FITC and 10 µL propidium iodide (PI) (Beyotime Institute of Biotechnology, Haimen, China) was added to staining. The staining process was undergone in dark at room temperature for 10–20 min. Then, fluorescence of FITC and PI was detected by FCM (FACSCalibur, BD Bioscience, Franklin Lakes, NJ, USA). Macquit software (BD Bioscience, Franklin Lakes, NJ, USA) was used to analyze the data.
Statistical analysis
All data were analyzed using SPSS 17.0 (IBM Analytics, New York, NY, USA) and GraphPad Prism 5. The quantitative date was showed as means ± standard deviation (SD). One-way ANOVA and t-test was used to analyze the difference. P<0.05 was defined as statistically significant.
Results
The expression of lncRNA DANCR is increased in ESCC tissues
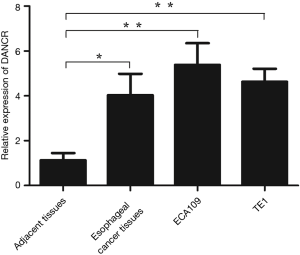
The expressions of lncRNA DANCR in ESCC tissues, adjacent tissues and ESCC cells were tested by qRT-PCR. The results of qRT-PCR analysis indicated that the relative expression of lncRNA DANCR was notably higher in ESCC tissues and ESCC cells than that in adjacent tissues (P=0.047, P<0.05). And the relative expression of DANCR in ECA109 cells was highest. As shown in Figure 1. So, the ECA109 cells were selected to use in following experiments.
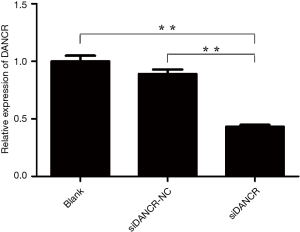
LncRNA DANCR promotes cell proliferation
In order to explore the biological effect of lncRNA DANCR on the ESCC cells, the ECA109 cell were transfected with siDANCR to knock down the expression of DANCR. Firstly, the expression changes of DANCR after transfection were analyzed by qRT-PCR. As shown in Figure 2, compared to the blank and siDANCR-NC groups, the expression of DANCR in the siDANCR transfection group was remarkably lower (P<0.01). The result suggested that the DANCR expression was significantly down-regulated after the successful siRNA transfection.
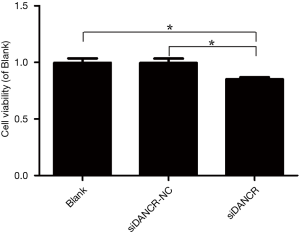
Secondly, we examined the influence of lncRNA DANCR on cell proliferation. MTT assay was performed to detect the effect of DANCR on the viability of ECA109 cells transfected with siDANCR. The viability in siDANCR transfection group approximately decreased 30%, compared to blank (P=0.044, P<0.05) or siDANCR-NC group (P=0.046, P<0.05). Meanwhile, significant difference between blank and siDANCR-NC groups did not exist (P>0.05). As shown in Figure 3, the result indicated that the viability of ECA109 cell could be inhibited by the down-regulated expression of DANCR.
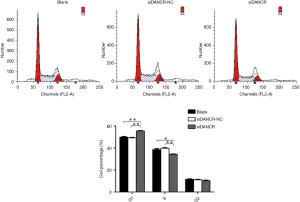
Next, the influence of DANCR on the cell cycle of ECA109 cells was further tested by FCM assay. The results of FCM assay revealed that the percent of S phase was significantly reduced in siDANCR transfection group, compared to blank or siDANCR-NC group (P<0.01). However, G1 phase was notably increased in siDANCR transfection group (P<0.01). As shown in Figure 4, notable difference of S phase, G1 phase and G2 phase between the blank and siDANCR-NC groups did not exist (P>0.05). The ECA109 cells treated with siDANCR were arrested in G1 phase, and knocking down DANCR could reduce the proliferation and the mitosis of ECA109 cells by approximately 15%.
LncRNA DANCR promotes cell migration and invasion
Cell migration and invasion is a central process in the evolution and progression of tumor. Wound healing assay and Transwell chamber were used to investigate the influence of DANCR on the cell migration and invasion of ECA109 cells. The cell motility of ECA109 cell treated with siDANCR transfection for 24 and 48 h were reduced 70.43% and 69.54% respectively, compared to blank or siDANCR-NC group (P<0.01) in wound healing assay. It was shown in Figure 5.
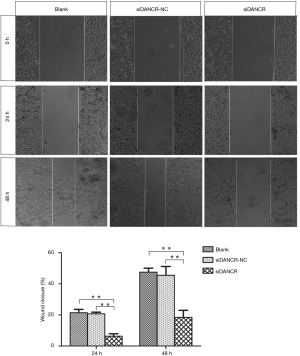
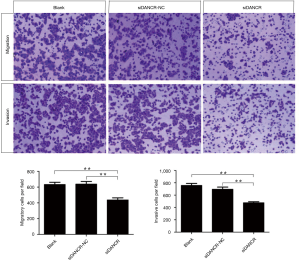
To further confirm the effect of DANCR on migration and invasion of ECA109 cells, Transwell chamber assays were undergone. The cells transferred from the upper chamber into the lower chamber indicated the migrated/invasive cells. The numbers of transferred cells in the DANCR interference group were notably fewer than that in the blank and siDANCR-NC groups (P<0.01) by approximately 30%. The results of Transwell assay were showed in Figure 6. Taken together, knock-down of DANCR could efficiently repress the migration and invasion of ECA109 cells.
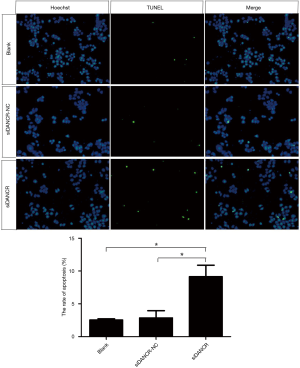
LncRNA DANCR inhibits cell apoptosis
The TUNEL immunofluorescence staining was used to determine the effect of DANCR on the apoptosis of ECA109 cells. The results demonstrated that the positive rate of TUNEL staining was approximately 10% in siDANCR transfection group, which was significantly higher than that of blank (P=0.022, P<0.05) and siDANCR-NC group (P=0.028, P<0.05). As shown in Figure 7. The result of TUNEL staining indicated that down-regulation DANCR promoted apoptosis of ECA109 cells.
Discussion
As one of the most endemic cancers in China, esophageal carcinoma has relatively poor prognosis with high relapse and invasion rates (24-26). Although the patients with esophageal cancer received continuous treatments like radiotherapy and chemotherapy, the 5-year recurrence rate of this disease remained high. It is not only the challenge but also the focus in cancer research now (27,28). Furthermore, gene therapy recently becomes a hot topic in cancer research (9,29). Researches about proliferation, invasion and migration that regulated by esophageal cancer related genes also attract lots of attention (30,31). In view of this phenomenon, explore new molecule targets on tumor is necessary (32-34).
LncRNAs, its transcripts over 200 nucleotides, had been proved to involve in multiple life proceed, such as transcriptional regulation and posttranscriptional regulation (35-37). The latest proofs showed that lncRNAs acted as vital parts in tumor growth or metastasis, and may work as predict markers (18,38). Jia et al. suggested that lncRNA DANCR could promote the invasion of prostate cancer by down-regulating TIMP2/3 (39). Previously, it was reported that lncRNA DANCR was up-regulated and increased the stem cell properties of liver cancer cells by the depression of CTNNB1 (23). Previous studies also showed that the down-regulation of DANCR suppressed the proliferation of many types of cancer cells including liver cancer, prostate cancer, colorectal cancer, etc., and induced differentiation or apoptosis of those tumor cells (23,40,41). So far, the expression and effect of lncRNA DANCR in ESCC on tumor growth and metastasis are still unknown. Additionally, identifying the molecular mechanisms of tumorigenesis is still a remarkable challenge.
Firstly, the expression of DANCR in ESCC tissues and cells was measured by qRT-PCR. Secondly, the biological roles of DANCR were investigated by various methods. We took ESCC cell line ECA109 cells as a research object. Our results revealed that the expression of lncRNA DANCR in ESCC tissues and cell lines was notably up-regulated, indicating that DANCR might take part in modulating the tumorigenesis and progression of ESCC.
The proliferation of cancer cells is the critical role during the development of esophageal carcinoma. It was discovered that DANCR expressed in esophageal cancer cells, and importantly, DANCR ectopic expression promoted proliferation and metastasis of ESCC. Migration and invasion of cancer cells result in a malignant phenotype. Our results showed that besides the down-regulation of DANCR significantly suppressed cell proliferation, and inhibited migration and invasion, and promoted cell apoptosis. However, the down-regulation of DANCR significantly promoted apoptosis.
Collectively, DANCR promotes the proliferation, invasion, and migration of ECA109 cells. It could work as a predict biomarker and potential therapeutic target in ESCC. But, there was a limitation that there was no data of in vivo examination and mechanisms in this study. Our results support the theory that silencing of DANCR could have potential beneficial effects on the prognostic and therapy for ESCC in the future.
Acknowledgements
Funding: This study was supported by grants from the National Natural Science Foundation of China (grant No. 81101159, 81502055), and the Natural Science Foundation of Jiangsu Province, China (grant No. BK20151268).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved ethically by the Committee of Affiliated Hospital of Nantong University (No. 2015-070). All patients were notified consent for the use of their tissues for scientific research.
References
- Morita FH, Bernardo WM, Ide E, et al. Narrow band imaging versus lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta-analysis. BMC Cancer 2017;17:54. [Crossref] [PubMed]
- Giacopuzzi S, Bencivenga M, Weindelmayer J, et al. Western strategy for EGJ carcinoma. Gastric Cancer 2017;20:60-8. [Crossref] [PubMed]
- Mannath J, Ragunath K. Role of endoscopy in early oesophageal cancer. Nat Rev Gastroenterol Hepatol 2016;13:720-30. [Crossref] [PubMed]
- Le Bras GF, Farooq MH, Falk GW, et al. Esophageal cancer: The latest on chemoprevention and state of the art therapies. Pharmacol Res 2016;113:236-44. [Crossref] [PubMed]
- Xiao F, Pu J, Wen Q, et al. Association between the ERCC2 Asp312Asn polymorphism and risk of cancer. Oncotarget 2017;8:48488-506. [Crossref] [PubMed]
- Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol 2017;27:215-21. [Crossref] [PubMed]
- Shi JF, Huang HY, Guo LW, et al. Quality-of-life and health utility scores for common cancers in China: a multicentre cross-sectional survey. Lancet 2016;388 Suppl 1:S29. [Crossref]
- Yue D, Zhang Z, Li J, et al. Transforming growth factor-beta1 promotes the migration and invasion of sphere-forming stem-like cell subpopulations in esophageal cancer. Exp Cell Res 2015;336:141-9. [Crossref] [PubMed]
- Chen GZ, Zhu HC, Dai WS, et al. The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J Thorac Dis 2017;9:849-59. [Crossref] [PubMed]
- Sohda M, Kuwano H. Current Status and Future Prospects for Esophageal Cancer Treatment. Ann Thorac Cardiovasc Surg 2017;23:1-11. [Crossref] [PubMed]
- Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med 2017;14:33-41. [Crossref] [PubMed]
- Myint ZW, Goel G. Role of modern immunotherapy in gastrointestinal malignancies: a review of current clinical progress. J Hematol Oncol 2017;10:86. [Crossref] [PubMed]
- Li X, Zhao L, Zhang W, et al. Prognostic value of supraclavicular nodes and upper abdominal nodes metastasis after definitive chemoradiotherapy for patients with thoracic esophageal squamous cell carcinoma. Oncotarget 2017;8:65171-85. [PubMed]
- Zhu Y, Li M, Bo C, et al. Prognostic Significance of and the Relationship Between Tumor-infiltrating and Circulating Lymphocytes to Macrophage/Monocyte Ratio in Patients With Esophageal Squamous Cell Cancer of Pathological Stage T3N0M0. Int J Radiat Oncol Biol Phys 2017;98:228.
- Schøler LV, Reinert T, Ørntoft MW, et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res 2017;23:5437-45. [Crossref] [PubMed]
- Dong X, Liao W, Zhang L, et al. RSPO2 suppresses colorectal cancer metastasis by counteracting the Wnt5a/Fzd7-driven noncanonical Wnt pathway. Cancer Lett 2017;402:153-65. [Crossref] [PubMed]
- Wang J, Li Q, Yuan J, et al. CDK4/6 inhibitor-SHR6390 exerts potent antitumor activity in esophageal squamous cell carcinoma by inhibiting phosphorylated Rb and inducing G1 cell cycle arrest. J Transl Med 2017;15:127. [Crossref] [PubMed]
- Peng F, Wang R, Zhang Y, et al. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer 2017;16:98. [Crossref] [PubMed]
- Zhang L, Yang C, Chen S, et al. Long Noncoding RNA DANCR Is a Positive Regulator of Proliferation and Chondrogenic Differentiation in Human Synovium-Derived Stem Cells. DNA Cell Biol 2017;36:136-42. [Crossref] [PubMed]
- Leone S, Bar D, Slabber CF, et al. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep 2017;18:1248-62. [Crossref] [PubMed]
- Chen X, Gao Y, Li D, et al. LncRNA-TP53TG1 participated in the stress response under glucose deprivation in glioma. J Cell Biochem 2017;118:4897-904. [Crossref] [PubMed]
- Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 2012;26:338-43. [Crossref] [PubMed]
- Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016;63:499-511. [Crossref] [PubMed]
- Zhan XH, Jiao JW, Zhang HF, et al. A three-gene signature from protein-protein interaction network of LOXL2- and actin-related proteins for esophageal squamous cell carcinoma prognosis. Cancer Med 2017;6:1707-19. [Crossref] [PubMed]
- Fu L, Qin YR, Ming XY, et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc Natl Acad Sci U S A 2017;114:E4631-40. [Crossref] [PubMed]
- Liu SY, Chen W, Chughtai EA, et al. PIK3CA gene mutations in Northwest Chinese esophageal squamous cell carcinoma. World J Gastroenterol 2017;23:2585-91. [Crossref] [PubMed]
- Liu Q, Cui X, Yu X, et al. Cripto-1 acts as a functional marker of cancer stem-like cells and predicts prognosis of the patients in esophageal squamous cell carcinoma. Mol Cancer 2017;16:81. [Crossref] [PubMed]
- Li LY, Xie YH, Xie YM, et al. Ezrin Ser66 phosphorylation regulates invasion and metastasis of esophageal squamous cell carcinoma cells by mediating filopodia formation. Int J Biochem Cell Biol 2017;88:162-71. [Crossref] [PubMed]
- Kiyohara MH, Dillard C, Tsui J, et al. EMP2 is a novel therapeutic target for endometrial cancer stem cells. Oncogene 2017;36:5793-807. [Crossref] [PubMed]
- Heczey A, Louis CU, Savoldo B, et al. CAR T Cells Administered in Combination with Lymphodepletion and PD-1 Inhibition to Patients with Neuroblastoma. Mol Ther 2017;25:2214-24. [Crossref] [PubMed]
- Chen L, Pan J. Dual cyclin-dependent kinase 4/6 inhibition by PD-0332991 induces apoptosis and senescence in oesophageal squamous cell carcinoma cells. Br J Pharmacol 2017;174:2427-43. [Crossref] [PubMed]
- Whiteside TL. Stimulatory role of exosomes in the context of therapeutic anti-cancer vaccines. Biotarget 2017;1:5. [Crossref]
- Prieur A, Cappellini M, Habif G, et al. Targeting the Wnt Pathway and Cancer Stem Cells with Anti-progastrin Humanized Antibodies as a Potential Treatment for K-RAS-Mutated Colorectal Cancer. Clin Cancer Res 2017;23:5267-80. [Crossref] [PubMed]
- Wu Y, Jiang M. The revolution of lung cancer treatment: from vaccines, to immune checkpoint inhibitors, to chimeric antigen receptor T therapy. Biotarget 2017;1:7. [Crossref]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Liang J, Xu L, Zhou F, et al. MALAT1/miR-127-5p regulates osteopontin (OPN)-mediated proliferation of human chondrocytes through PI3K/Akt pathway. J Cell Biochem 2018;119:431-9. [Crossref] [PubMed]
- Fang Q, Xu T, Wu C, et al. Biotargets in neural regeneration. Biotarget 2017;1:6. [Crossref]
- Deng B, Cheng X, Li H, et al. Microarray expression profiling in the denervated hippocampus identifies long noncoding RNAs functionally involved in neurogenesis. BMC Mol Biol 2017;18:15. [Crossref] [PubMed]
- Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget 2016;7:37868-81. [Crossref] [PubMed]
- Ma X, Wang X, Yang C, et al. DANCR Acts as a Diagnostic Biomarker and Promotes Tumor Growth and Metastasis in Hepatocellular Carcinoma. Anticancer Res 2016;36:6389-98. [Crossref] [PubMed]
- Liu Y, Zhang M, Liang L, et al. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 2015;8:11480-4. [PubMed]


