
Editorial comments for the circular RNA circPRKCI promotion of tumor growth in lung adenocarcinoma
Lung cancer is one of the leading cause of death worldwide (1). There are two major forms of lung cancer: non-small cell lung cancer (NSCLC) (about 85%) and small cell lung cancer (SCLC) (about 15%). Treatments for lung cancer are mainly surgical resection, chemotherapy and targeted therapy, however, these treatments rarely achieve a cure, and the 5-year survival rate is still low (2). Therefore, there is an urgent need for new diagnostic biomarkers to ensure that lung cancer patients can be treated in an effective manner and survive longer. The molecular origins and evolution of the lung cancer is showing a stepwise continuum most notably the progression of atypical adenomatous hyperplasia (AAH) to adenocarcinoma in situ (AIS) and to minimally invasive adenocarcinoma (MIA). The morphological appearance of the tumor and molecular processes which are involved in tumor progression are both supported by this model. Based on the WHO classification and according to the extent of invasiveness, adenocarcinoma is divided into AIS (preinvasive lesion), MIA, and overt invasive adenocarcinoma (3).
AAH has been recognized as an early lesion in the process leading to lung adenocarcinoma (LAC). A variety of molecular findings support this sequential model. AAH demonstrates several molecular features like; increased expression of CCND1 (cyclin D1), and decreased expression of CDKN2A (p16) (4,5), mutations in genes EGFR, BRAF, KRAS, FGFR3, and ERBB2 (6), and loss of heterozygosity of chromosomes, most notably 9q and 16p (7). Clonal expansion in AAH was also described (8,9). On the other hand, AIS is an intermediate step between AAH and MIA (3). Some specific DNA hyper methylation changes were detected in AIS but not in AAH (10). Telomere shortening and DNA damage response (DDRs) are considered as early events in lung carcinogenesis (11).
MIA is an early invasive adenocarcinoma. In the transition from AIS to MIA EGFR amplification was found as an independent event of the initiating early mutation in EGFR and KRAS genes (12). Repression of TGFBR2 is considered to contribute to invasiveness (13) and the amplification and overexpression of PDCD6 and TERT on chromosome 5p promote invasion (14). However, features besides amplification or overexpression of oncogenic proteins and deletion or repression of tumor suppressor are involved in tumorigenesis.
With the advent of next generation sequencing technologies (NGS), circular RNAs (circRNAs) were identified as a promising biomarker for a variety of cancers including lung cancer. They show cell phenotype specificity in a variety of tissues and are quite stable. CircRNAs can act as miRNA sponges and can regulate the transcription of parental genes. CircRNAs may vary in length significantly, most of them being longer than 200 nt (15). Current knowledge about the involvement of circRNAs in cancer development and progression is limited, and the role of circRNAs as miRNA sponges has been proposed as the most frequent mechanism of circRNA activity in tumor cells. Generally, miRNAs are included in various cell processes, including cellular differentiation, development, proliferation, and apoptosis, where they play an important role as regulators of gene expression. These miRNA-mediated processes are frequently deregulated in cancer and can contribute to cancer initiation and progression. Since many circRNAs regulate miRNA action through binding, dysregulation of circRNA expression may affect their interaction with tumor-associated miRNAs, indicating an important role of circRNAs in regulating cancer (16,17).
Several lung cancer circRNAs expression studies revealed that some circRNAs play an important regulatory role in the lung carcinogenesis (15). Especially, identification of circRNAs involved in the earliest steps of cancer progression could have an important therapeutic potential.
In a cohort of an early LAC Zhao et al. performed circRNA microarray analysis to investigate the circRNAs expression signature in tumor tissues and normal tissue near tumors (18). They found 357 circRNAs differentially expressed suggesting their potential roles in lung cancer. To confirm the microarray data, selected circRNAs were detected and verified in tissues or cell lines by the qRT-PCR methods. They found that the expression levels of hsa_circRNA_404833 and hsa_circRNA_406483 were upregulated in LAC but were almost undetectable in the adjacent normal tissues. Furthermore, the circRNA/miRNA interaction algorithm predicted that hsa_circRNA_404833 is potentially able to interact with miR-149-5p. miR-149-5p is involved in the acquired gefitinib (EGFR inhibitor) resistance in LAC (18).
In a similar study Zhu et al. (19) described the expression profiling of circRNAs in LAC patients using tissue samples and plasma samples. qPCR validation and the ROC analyses revealed that hsa_circ_0013958 had a high degree of specificity and sensitivity. Additional results showed that hsa_circ_0013958 increased cell proliferation and invasion at the same time as it inhibited cell apoptosis. Hsa_circ_0013958 also acted to sequester miR-134 and thus up-regulate oncogenic CCND1, which plays a pivotal role in the development of NSCLC (19).
Hsa_circ_0012673 is overexpressed in LACs and promotes cell proliferation through the miR-22/ErbB3 pathway. Knocking down hsa_circ_0012673 or silencing miR-22 promotes proliferation of LAC. Interestingly the expression of ERBB3 in tumor tissues of LAC patients when upregulated positively correlated with hsa_circ_0012673, but negatively correlated with miR-22 expression what was confirmed also by Western blot (20).
Li et al. (21) identified a novel circRNA, hsa_circ_0000792, which was significantly upregulated and may play an important role in LAC. A network of circRNA-miRNA interactions was constructed and miR-375 interacted with most nodes. They proposed a mechanism in which hsa_circ_0000792 competitively binds with miR-375 and revoke the inhibiting effect on their associated target genes (22).
In most cases CircRNAs are mainly derived from the exons of protein-coding genes, although they can also arise from intronic, intergenic, UTR regions and ncRNA loci (23).
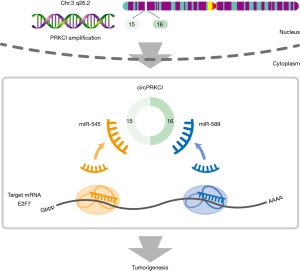
CircRNAs arising from copy number variation (CNV) regions have not been identified before. CNV is a form of genomic structural variation leading to gains and losses of DNA segments. Due to genomic instability, somatic CNVs are extremely common and play a significant role during the development of cancer, where proto-oncogenic genes are usually amplified, and tumor suppressors are deleted. In their work Qiu et al. (24) further explored the hidden noncoding transcripts within recurrent CNV loci in cancer. Bioinformatics analyses of circRNAs and focal CNV in LAC revealed circular RNA (circPRKCI) from the genomic 3q26.2 region. Indeed, in this region there are most frequent genomic aberrations in multiple cancers. In further experimental work they proved that circPRKCI could function as a sponge for two miRNAs namely miR-545 and miR-589. As a consequence expression levels of E2F7 are increased promoting the tumorigenesis of LAC (24).
In their comprehensive work, Qiu et al. first performed microarray analysis to characterize the expression signatures of circRNAs and mRNAs in LAC patients. They found 107 circRNAs and 1691 mRNAs to be differentially expressed. However, the expression levels of circRNAs and mRNA levels of their host genes were not in correlation. Detailed analysis of upregulated circRNA in the amplified regions identified also two novel amplified host genes (PRKCI and P4HB).
CircPRKCI located at 3q26.2 derive from back-splicing of two exons (exons 15 and 16) in the PRKCI gene. Further characterization of circPRKCI with PCR confirmed its existence in cDNA. qRT-PCR results showed highly up-regulated circPRKCI in LAC and its positive correlation with TNM stage. CircPRKCI expression was confirmed in lung cancer tissues and normal tissue near tumors by chromogenic in situ hybridization (CISH). Tissue microarray (TMA) was used in the experiment. A strong positive correlation between circPRKCI expression and TNM stage was found. High circPRKCI levels could be used as an independent poor prognostic factor for LAC patients. Partial intercorrelation between PRKCI CNV and circPRKCI expression was also established.
To investigate the biological function of circPRKCI in vitro they performed siRNA and knockdown experiments. Knockdown of circPRKCI significantly weaken cell proliferation and arrested cells in G1 but did not affect cell apoptosis while overexpression of circPRKCI promoted proliferation and migration of LAC cells. In LAC cell lines si-circPRKCI treatment significantly weakened the invasion capacity of the cells.
The question, which arises, is the mechanism of generation of such high levels of circPRKCI. They found that more than 90% of circPRKCI was present in the cytoplasm in LAC cell lines.
To further delineate the molecular mechanism of circPRKCI RNA, an immunoprecipitation (RIP) assay was conducted. RNA transcripts that bind to Ago2 in LAC (SPC-A1 and A549) cells was pulled down. Results showed that endogenous circPRKCI was effectively pulled down by anti-Ago2. To determine if circPRKCI could sequester miRNAs, they performed a miRNA pull-down assay and found that circPRKCI was efficiently enriched by miR-589 and miR-545. RIP assays further revealed that miR-545 and miR-589 were efficiently pulled down by the anti-Ago2 antibody. Additional experiments indicated that circPRKCI functions as a miRNA sponge not affecting the expression of miR-545 and miR-589, which act as inhibitors of the proliferation of LAC cells and inducers of G1 phase.
Both miRNAs are involved in LAC cells tumorigenesis suppression. In the ceRNA theory, circPRKCI expression should be in correlation with its target genes. Indeed, they identified seven candidate target genes from microarray data and found that only one of them, E2F7 was down-regulated after silencing of circPRKCI. Using miRNA biotin pull-down assay, cloning of wild-type and mutant 3’-UTR of E2F7 mRNA and a dual luciferase reporter assay they verified that E2F7 mRNA was a direct target of miR-545 and miR-589. In addition, they found that expression levels of both miR-545 and miR-589 directly affect the level of E2F7 via downregulation mechanism. Silencing of E2F7 inhibited LAC cell proliferation.
E2F7 is a transcription factor that plays a key role in the timely expression of genes required for cell cycle progression. E2F7 downregulates p21 leading to upregulation of Cyclin D1 (25).
Further experiments confirmed that silencing circPRKCI indeed decreased the levels of E2F7 and vice versa. As a consequence of decreased E2F7 in downstream pathway also levels of p21 and Cyclin D1 were altered. The in vitro experiments showed that circPRKCI operates via circPRKCI-miR-545/589-E2F7 axis and promotes LAC cell proliferation (24).
To explore in vivo function of circPRKCI, both a xenograft tumor model in nude mice and female LAC patient-derived tumor xenografts (PDTXs) were established. Cells transfected with si-circPRKCI produced smaller tumors in comparison with siRNA control transfected cells.
The therapeutic potential of circPRKCI was evaluated in PDTXs by intratumoral injection of cholesterol-coupled si-circPRKCI and a control siRNA. The results demonstrated that si-circPRKCI distinctly inhibited growth of PDTX in vivo. In both in vivo experiments was therefore shown that circPRKCI could serve as a possible therapeutic target of LAC.
EGFR tyrosine kinase inhibitors (EGFR-TKIs) are often the first-line therapy in EGFR mutation-positive LAC patients. In order to demonstrate whether circPRKCI could alter the therapeutic potential of EGFR-TKI proliferation assays in EGFR-TKI sensitive cells was performed. Combination of gefitinib with si-circPRKCI was more effective than gefitinib or si-circPRKCI alone, which again indicated circPRKCI as a promising therapeutic target.
In summary, Qiu et al. (24) identified a novel circPRKCI, which is overexpressed in LAC tissues. They also established the mechanism for tumor proliferation via miR-545 and miR-589 binding and E2F7 inhibition (Figure 1). This novel circular RNAs research provides circPRKCI a potential target for LAC patients treatment.
The discovery of LAC-related circRNAs thus offer new opportunities for the diagnosis and treatment of lung cancer. The comprehensive study of Qiu et al. (24) has initiated the opportunity for the discovery of other circRNAs transcribed from CNVs with similar prognostic and therapeutic potential as circPRKCI. Further discoveries of the biological functions of circRNAs and their regulatory mechanisms in cancer are likely to show also novel diagnostic and treatment possibilities for cancer.
Acknowledgements
We would like to acknowledge Dr. Michael Dean for proofreading of our editorial and Matjaž Lužar for drawing Figure 1.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Wu K, House L, Liu W, et al. Personalized targeted therapy for lung cancer. Int J Mol Sci 2012;13:11471-96. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Tominaga M, Sueoka N, Irie K, et al. Detection and discrimination of preneoplastic and early stages of lung adenocarcinoma using hnRNP B1 combined with the cell cycle-related markers p16, cyclin D1, and Ki-67. Lung Cancer 2003;40:45-53. [Crossref] [PubMed]
- Licchesi JD, Westra WH, Hooker CM, et al. Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res 2008;14:2570-8. [Crossref] [PubMed]
- Xu X, Li N, Zhao R, et al. Targeted next-generation sequencing for analyzing the genetic alterations in atypical adenomatous hyperplasia and adenocarcinoma in situ. J Cancer Res Clin Oncol 2017;143:2447-53. [Crossref] [PubMed]
- Takamochi K, Ogura T, Suzuki K, et al. Loss of heterozygosity on chromosomes 9q and 16p in atypical adenomatous hyperplasia concomitant with adenocarcinoma of the lung. Am J Pathol 2001;159:1941-8. [Crossref] [PubMed]
- Nakayama H, Noguchi M, Tsuchiya R, et al. Clonal growth of atypical adenomatous hyperplasia of the lung: cytofluorometric analysis of nuclear DNA content. Mod Pathol 1990;3:314-20. [PubMed]
- Niho S, Yokose T, Suzuki K, et al. Monoclonality of atypical adenomatous hyperplasia of the lung. Am J Pathol 1999;154:249-54. [Crossref] [PubMed]
- Selamat SA, Galler JS, Joshi AD, et al. DNA methylation changes in atypical adenomatous hyperplasia, adenocarcinoma in situ, and lung adenocarcinoma. PLoS One 2011;6:e21443. [Crossref] [PubMed]
- Nakanishi K, Kawai T, Kumaki F, et al. Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin Cancer Res 2003;9:1105-11. [PubMed]
- Soh J, Toyooka S, Ichihara S, et al. Sequential molecular changes during multistage pathogenesis of small peripheral adenocarcinomas of the lung. J Thorac Oncol 2008;3:340-7. [Crossref] [PubMed]
- Borczuk AC, Kim HK, Yegen HA, et al. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am J Respir Crit Care Med 2005;172:729-37. [Crossref] [PubMed]
- Aviel-Ronen S, Coe BP, Lau SK, et al. Genomic markers for malignant progression in pulmonary adenocarcinoma with bronchioloalveolar features. Proc Natl Acad Sci U S A 2008;105:10155-60. [Crossref] [PubMed]
- Chen Y, Wei S, Wang X, et al. Progress in research on the role of circular RNAs in lung cancer. World J Surg Oncol 2018;16:215. [Crossref] [PubMed]
- Chen D, Ma W, Ke Z, et al. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 2018;17:2080-90. [Crossref] [PubMed]
- Bolha L, Ravnik-Glavac M, Glavac D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int J Genomics 2017;2017:6218353. [Crossref] [PubMed]
- Zhao J, Li L, Wang Q, et al. CircRNA Expression Profile in Early-Stage Lung Adenocarcinoma Patients. Cell Physiol Biochem 2017;44:2138-46. [Crossref] [PubMed]
- Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J 2017;284:2170-82. [Crossref] [PubMed]
- Wang X, Zhu X, Zhang H, et al. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun 2018;496:1069-75. [Crossref] [PubMed]
- Li Y, Hu J, Li L, et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun 2018;503:2089-94. [Crossref] [PubMed]
- Li S, Sun X, Miao S, et al. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer 2018;9:924-30. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Qiu M, Xia W, Chen R, et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res 2018;78:2839-51. [Crossref] [PubMed]
- Mitxelena J, Apraiz A, Vallejo-Rodriguez J, et al. E2F7 regulates transcription and maturation of multiple microRNAs to restrain cell proliferation. Nucleic Acids Res 2016;44:5557-70. [Crossref] [PubMed]


