Rapid detection of Streptococcus pneumoniae by real-time fluorescence loop-mediated isothermal amplification
Introduction
Streptococcus pneumoniae is a Gram-positive, alpha-hemolytic, aero tolerant anaerobic member of the genus Streptococcus (1). A significant human pathogenic bacterium, S. pneumoniae was recognized as a major cause of pneumonia in the late 19th century, and is the subject of many humoral immunity studies (2).
S. pneumoniae resides asymptomatically in the nasopharynx of healthy carriers. However, in susceptible individuals, such as elderly and immunocompromised people and children, the pathogen can spread to other locations and cause disease. S. pneumoniae is the main cause of community acquired pneumonia and meningitis in children and the elderly, and of septicemia n HIV-infected persons (3).
Streptococcus pneumoniae is a major cause of diseases such as pneumonia, meningitis and sepsis, though each of these diseases is also caused by other organisms. In the developed world, serious disease occurs mainly in children below 2 years of age and in the elderly. In developing countries, the disease is common in children under 2 years, including newborn infants; rates of the disease in the elderly population are largely unknown (4).
Diagnosis is generally made based on clinical suspicion along with a positive culture from a sample from virtually any place in the body. An ASO titre of >200 units is significant. S. pneumoniae is, in general, optochin sensitive, although optochin resistance has been observed (5).
The microbiological diagnosis made microscopically and culturally. Pneumococci can be well on blood agar plates or in liquid culture media containing blood cultured. However, the period between sample collection and processing be as short as possible, because the cause of the pneumococcal autolytic enzyme system for the rapid death of the pathogen (6).
LAMP is a relatively new DNA amplification technique, which due to its simplicity, ruggedness, and low cost could provide major advantages. In LAMP, the target sequence is amplified at a constant temperature of 60-65 °C using either two or three sets of primers and a polymerase with high strand displacement activity in addition to a replication activity. Typically, four different primers are used to identify six distinct regions on the target gene, which adds highly to the specificity (7). Due to the specific nature of the action of these primers, the amount of DNA produced in LAMP is considerably higher than PCR based amplification. The corresponding release of pyrophosphate results in visible turbidity due to precipitation, which allows easy visualization by the naked eye, especially for larger reaction volumes or via simple detection approaches for smaller volumes (8). The reaction can be followed in real-time either by measuring the turbidity or the signals from DNA produced via fluorescent dyes that intercalate or directly label the DNA, and in turn can be correlated to the number of copies initially present. Hence, LAMP can also be quantitative (9). While LAMP is widely being studied for detecting infectious diseases such as tuberculosis, malaria, and sleeping sickness in developing regions, it has yet to be extensively validated for other common pathogens. Our study is to establish a real time LAMP for the diagnosis of Streptococcus pneumoniae.
Methods
Bacterial strains
Bacterial strains were isolated from human clinical samples which were collected following the third affiliated Hospital of Guangzhou Medical University approved procedures. The study was approved by the Ethics Committee of the Third affiliated Hospital of Guangzhou Medical University and all aspects of the study comply with the Declaration of Helsinki.
Streptococcus pneumoniae ATCC 49619
Streptococcus pneumoniae ATCC 49619 was kindly provided by Professor Jia-Yun Liu of the Fourth Military Medical University. The genomic DNA was subjected to serial 10-fold dilutions in sterilized distilled water to produce concentrations ranging from 0.3 ng/uL to 3,000 ng/mL and assess the correlation between time to amplification and amount of target DNA. In addition, to evaluate the specificity of real-amp, DNA extracts from the array of bacterial species were tested.
DNA extraction
DNA was isolated from all the samples using a QIAamp DNA Mini Kit [Qiagen, Valencia, CA (Qiagen method)]. Briefly, each sputum sample was liquefied in an equal volume of sputasol solution, placed 60 minutes at 37 °C and incubate until liquifaction is complete. One milliliter of the liquefaction of sputum was moved into a 1.5 mL eppendorf tube, and then centrifuged at 12,000 rpm for 10 minutes and the precipitate collected. The precipitate was washed by one milliliter of 16 TE (Tris-EDTA) buffer once, and then centrifuged at 12,000 rpm for 10 minutes and the precipitate collected. Forty microliters of sterilized distilled water was added to the tube containing the precipitate and mixed. The tube was heated on a heat-block at 100 °C for 15 minutes, and then placed on ice for 10 minutes. Finally, the tube was then centrifuged at 12,000 rpm for 10 minutes and the supernatants were collected and used in the real-amp assays.
Design of real-amp primers
The primers forward outer primer (F3), backward outer primer (B3), forward internal primer (FIP), backward internal primer (BIP) and loop backward primer (LB) listed in Table 1 for the real-amp test were designed by targeting the conserved regions of the pneumolysin a gene of Streptococcus pneumonia.
Real-amp method
The real-amp method was performed using the commercially available DNA thermostatic amplification kit (Guangzhou Diao Bio-technology Co., Ltd., Guangdong, China) following the manufacturer’s instructions. Reactions were performed in 25 mL total volume containing 26 reaction buffer (40 mm Tris-HCl PH 8.8, 20 mm KCl, 16 mm MgSO4, 20 mm (NH4)2SO4, 0.2% Tween-20, 0.8M Betaine, 2.8 mm of dNTPs each), 0.5 mL of a 1:100 dilution SYBR green I (Invitrogen), 0.2 mm of each outer primers of F3 and B3, 1.6 mm of each inner primers of FIP and BIP, 0.8 mm of loop primer of LB, and 8 units of Bst polymerase (New England Biolabs, Ipswich, MA). DNA amplification was carried out at 63 °C for 60 minutes using the ESE-Quant Tube scanner which was set to collect fluorescence signals. The ESE-Quant Tube scanner used in this study was developed by a company (QIAGEN Lake Constance GmbH, Stockach, Germany). This device has an eight tube holder heating block with adjustable temperature settings and spectral devices to detect amplified product using fluorescence spectra. The unit is completely portable and can be operated with a Li-Ion rechargeable power pack without external power supply. A small liquid crystal display (monitor) is available to display the results (as positive or negative) without the need of a computer. However, the device can also be used together with a computer to generate real time amplification plots as the reaction progresses. In this study, each sample was tested three times.
Results
Real-amp primers designed in this study
The target selection for primer design can be accomplished by using the Primer Explorer (http://Primer Explorer.jp/e/v4manual/Index.html). The primer specificity was checked using the basic local alignment search tool (BLAST) against human DNA and other Streptococcus sequences in the nonredundant GenBank database. And the primers used in this study listed in Table 1.

Full table
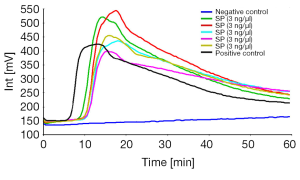
We were able to amplify Streptococcus pneumoniae ATCC 49619 within 60 minutes without loop backward primer. The fluorescence peak typically persisted for about 20 minutes (Figure 1A,B). And when we amplify Streptococcus pneumoniae with loop backward primer, the amplification time shorten to 20 minutes, and the fluorescence peak typically persisted for about 10 minutes. No amplification was seen with negative control. The fluorescence in millivolts of Strep-2 is higher than Strep-1, so we chose Strep-2 for the next experiments (Figure 1C,D).
Sensitivity of the real-amp method
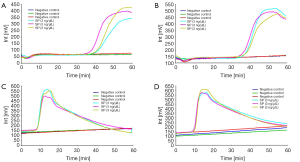
The limits of detection of real-amp were determined using DNA obtained from Streptococcus pneumoniae ATCC 49619. The DNA was diluted from 0.3 or 3 to 3,000 pg/uL. This assay required at least 300 to 3,000 pg/uL for the detection of Streptococcus pneumoniae ATCC 49619. The fluorescence peak typically persisted for about 10 minutes (Figure 2A). More time for amplification was required for samples with lower DNA concentration although no clear correlation was observed between time to amplification and the DNA concentration.
Specificity of the real-amp method
Clinical sputum samples were first identificated by VITEK 2 system, and then assessed by real-amp established in this study.
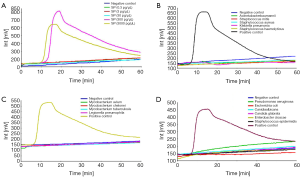
We selected 15 species belong to other genera. The results showed that the Real-Amp assay could effectively differentiate Streptococcus pneumoniae from 15 strains of other non-Streptococcus pneumoniae (Figure 2B-D).
Of the 15 species tested in VITEK 2 system, all samples were confirmed to be negative in the real-amp assay. Figure 2B shows that Acinetobacter baumannii, Streptococcus mitis, Staphylococcus aureus, Klebiella pneumonia and Staphylococcus haemolyticus were confirmed negative by real-amp assay. Figure 2C shows that Mycobacterium avium, Mycobacterium chelonei, Mycobacterium tuberculosis and Legionella pneumophila were confirmed negative by real-amp assay. Figure 2D shows that Pseudomonas aeruginosa, Escherichia coli, Canidia albicans, Candida glabrata, Enterobacter cloacae and Staphylococcus epidermidis confirmed negative by real-amp assay.
Repeatability of the real-amp method
The results showed that the repeatability of the real-amp method is good (Figure 3).
Discussion
In recent years, molecular biology, especially LAMP, has become the most valuable technology for clinical microbiology diagnosis (10-13). LAMP is an autocycling and strand displacement DNA synthesis method involving the use of the large fragment of Bst DNA polymerase and two pairs of specific inner and outer annular primers designed based on the gene sequence of different purposes. In the LAMP reaction, the design of the inner primer is capable of hybridizing to the DNA region in the target sequence, synthetizing the complementary strand, and producing a type of dumbbell-shaped DNA (14). Then this special structure use itself as a template for DNA synthesis to convert stem-loop DNA, which is the starting configuration of the LAMP cycling reaction. Because the inner primer hybridization in the ring of the stem-loop structure, so after the replacement the primer can produce a gaped stem-loop-like DNA which may be attaching target sequence (15). In the LAMP reaction process, pyrophosphate ions generated from the DNTP may bind to Mg2+ in a reaction solution, and became ivory precipitation magnesium pyrophosphate. The results also can be determined under a naked eye by adding fluorescent dye. LAMP reaction can be processed at a constant temperature by using simple devices with no need for temperature cycle, and its rapid and simple features give it an advantage over PCR (16-18).
LAMP is a single tube technique for the amplification of DNA. LAMP is isothermal nucleic acid amplification. Isothermal amplification in general obviates the need for thermal cyclers (10,19,20). Detection of amplification product can be determined via photometry for turbidity caused by an increasing quantity of magnesium pyrophosphate in solution as a byproduct of amplification or with addition of SYBR green, a color change can be seen without equipment. Also in-tube detection of DNA amplification is possible using manganese loaded calcium which starts fluorescing upon compellation of manganese by pyrophosphate during in vitro DNA synthesis (21-23).
LAMP has the potential to be used as a simple screening assay in the field or at the point of care by clinicians (16-18,24). As previously mentioned, LAMP is isothermal which eradicates the need for expensive thermo cyclers used in conventional PCR, it may be a particularly useful method for infectious disease diagnosis in low and middle income countries (10,19,20,25,26).
In a conclusion, LAMP is easy to operate and no need of complex instruments. In conclusion, as a quick and easy detection of Streptococcus pneumoniae, real-time Fluorescence Loop-Mediated Isothermal Amplification was successfully established, laid the foundation for the diagnosis and treatment of Streptococcus pneumonia.
Acknowledgements
Funding: This study was supported by grants from the Guangzhou Medical University Youth Fund Projects [grant number 2010A26] and the Medical and Health Science and Technology Projects of Guangzhou City [grant number 20131A011157].
Disclosure: The authors declare no conflict of interest.
References
- Uzuner A, Ilki A, Akman M, et al. Nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae in healthy children. Turk J Pediatr 2007;49:370-8. [PubMed]
- Abut LI, Apan T, Otlu B, et al. The characteristics of nasopharyngeal Streptococcus pneumoniae in children attending a daycare unit. New Microbiol 2008;31:357-62. [PubMed]
- Adeleye A, Uju L, Idika N, et al. Cotrimoxazole resistance in Streptococcus pneumoniae isolated from sputum of HIV-positive patients. West Indian Med J 2008;57:497-9. [PubMed]
- Aguiar SI, Serrano I, Pinto FR, et al. Changes in Streptococcus pneumoniae serotypes causing invasive disease with non-universal vaccination coverage of the seven-valent conjugate vaccine. Clin Microbiol Infect 2008;14:835-43. [PubMed]
- Antonio M, Hakeem I, Awine T, et al. Seasonality and outbreak of a predominant Streptococcus pneumoniae serotype 1 clone from The Gambia: expansion of ST217 hypervirulent clonal complex in West Africa. BMC Microbiol 2008;8:198. [PubMed]
- Azzari C, Moriondo M, Indolfi G, et al. Molecular detection methods and serotyping performed directly on clinical samples improve diagnostic sensitivity and reveal increased incidence of invasive disease by Streptococcus pneumoniae in Italian children. J Med Microbiol 2008;57:1205-12. [PubMed]
- Aoi Y, Hosogai M, Tsuneda S. Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. J Biotechnol 2006;125:484-91. [PubMed]
- Fukuda S, Takao S, Kuwayama M, et al. Rapid detection of norovirus from fecal specimens by real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol 2006;44:1376-81. [PubMed]
- Kouguchi Y, Fujiwara T, Teramoto M, et al. Homogenous, real-time duplex loop-mediated isothermal amplification using a single fluorophore-labeled primer and an intercalator dye: Its application to the simultaneous detection of Shiga toxin genes 1 and 2 in Shiga toxigenic Escherichia coli isolates. Mol Cell Probes 2010;24:190-5. [PubMed]
- Soleimani M, Shams S, Majidzadeh-A K. Developing a real-time quantitative loop-mediated isothermal amplification assay as a rapid and accurate method for detection of Brucellosis. J Appl Microbiol 2013;115:828-34. [PubMed]
- Lin Z, Zhang Y, Zhang H, et al. Comparison of loop-mediated isothermal amplification (LAMP) and real-time PCR method targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Vet Parasitol 2012;185:296-300. [PubMed]
- Parida M, Shukla J, Sharma S, et al. Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for rapid and real-time detection of the swine-origin influenza A H1N1 virus. J Mol Diagn 2011;13:100-7. [PubMed]
- Lucchi NW, Demas A, Narayanan J, et al. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One 2010;5:e13733. [PubMed]
- Yang B, Wang X, Li H, et al. Comparison of loop-mediated isothermal amplification and real-time PCR for the diagnosis of tuberculous pleurisy. Lett Appl Microbiol 2011;53:525-31. [PubMed]
- Boyanton BL Jr, Sural P, Loomis CR, et al. Loop-mediated isothermal amplification compared to real-time PCR and enzyme immunoassay for toxigenic Clostridium difficile detection. J Clin Microbiol 2012;50:640-5. [PubMed]
- Xu H, Zhang L, Shen G, et al. Establishment of a novel one-step reverse transcription loop-mediated isothermal amplification assay for rapid identification of RNA from the severe fever with thrombocytopenia syndrome virus. J Virol Methods 2013;194:21-5. [PubMed]
- Xie J, Liu G, Tian Z, et al. Development of loop-mediated isothermal amplification (LAMP) for detection of Theileria equi. Acta Trop 2013;127:245-50. [PubMed]
- Wang Q, Zhou Y, Li S, et al. Real-Time Fluorescence Loop Mediated Isothermal Amplification for the Detection of Acinetobacter baumannii. PLoS One 2013;8:e66406. [PubMed]
- Singh R, Savargaonkar D, Bhatt R, et al. Rapid detection of Plasmodium vivax in saliva and blood using loop mediated isothermal amplification (LAMP) assay. J Infect 2013;67:245-7. [PubMed]
- Nie K, Zhao X, Ding X, et al. Visual detection of human infection with influenza A (H7N9) virus by subtype-specific reverse transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. Clin Microbiol Infect 2013;19:E372-5. [PubMed]
- Zhao X, Li Y, Park M, et al. Loop-mediated isothermal amplification assay targeting the femA gene for rapid detection of Staphylococcus aureus from clinical and food samples. J Microbiol Biotechnol 2013;23:246-50. [PubMed]
- Zhao F, Liu Z, Gu Y, et al. Detection of Mycoplasma pneumoniae by colorimetric loop-mediated isothermal amplification. Acta Microbiol Immunol Hung 2013;60:1-9. [PubMed]
- Xue-han Z, Qing Y, Ya-dong L, et al. Development of a LAMP for Rapid Detection of Different Intimin Variants from Attaching and Effacing Microbial Pathogens. J Med Microbiol 2013;62:1665-72. [PubMed]
- Zhang J, Zhu J, Ren H, et al. Rapid visual detection of highly pathogenic Streptococcus suis serotype 2 using loop-mediated isothermal amplification. J Clin Microbiol 2013;51:3250-6. [PubMed]
- Tsai MA, Wang PC, Yoshida T, et al. Development of a sensitive and specific LAMP PCR assay for detection of fish pathogen Lactococcus garvieae. Dis Aquat Organ 2013;102:225-35. [PubMed]
- Kaewphinit T, Arunrut N, Kiatpathomchai W, et al. Detection of Mycobacterium tuberculosis by using loop-mediated isothermal amplification combined with a lateral flow dipstick in clinical samples. Biomed Res Int 2013;2013:926230.