
Is SARS-COV-2 associated with alpha-1 antitrypsin deficiency?
Introduction
Since the outbreak of a novel coronavirus (SARS-CoV-2), over 619 million of cases have been reported, and coronavirus disease (COVID) has killed almost 6.6 million people [World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports] (1-3). COVID-19 is associated with a striking variety of clinical manifestations ranging from mild (80%) to severe forms (ICU admission, orotracheal intubation or death) (4) that may cause severe acute respiratory distress, or death (5).
The likeliness of a poor course (admission to the intensive care unit, orotracheal intubation or death) has been associated with various risk factors (4). Therefore, measures are necessary to prevent complications and mortality (6). In this line, some recent studies suggest that patients with alpha-1 antitrypsin (A1AT) deficiency are at an increased risk of contracting SARS-CoV-2 infection (7,8). The A1AT protease inhibitor has been suggested to potentially prevent SARS-CoV-2 entry into host cells through the inhibition of S protein priming by TMPRSS2 (9). Hence, it is plausible to think that A1AT deficiency may play a role in severe forms of COVID-19.
This paper is aimed at assessing the relationship between S and/or Z alleles in heterozygous or homozygous genotypes and the risk for SARS-COV-2 infection, and evaluating the association of A1AT genotype and disease severity in subjects with European ancestry.
Methods
The association between A1AT deficiency and SARS-COV-2 was explored using data from the Spanish COalition to Unlock Research on host GEnetics (SCOURGE) study on COVID-19 (https://www.scourge-covid.org) (10), which included 11,939 COVID-19 patients treated in 34 hospitals and research centers of Spain between March and December 2020. Clinical data was entered into the Research Electronic Data Capture (REDCap) database and classified according to five levels of severity {asymptomatic; mild (with symptoms but without pulmonary infiltrates or need for oxygen therapy); moderate (with pulmonary infiltrates affecting <50% of the lungs or need for supplemental oxygen therapy); severe (hospitalized patients meeting any of the following criteria: PaO2 <65 mmHg or SaO2 <90%; PaO2/FiO2 <300, SaO2/FiO2 <440; dyspnea; respiratory frequency ≥22 bpm; or infiltrates affecting >50% of the lungs); and critical [admission to the intensive care unit or need for mechanical ventilation (invasive or non-invasive)]} (11). REDCap is a secure web application designed to support data capture for research studies that enables the collection and management of demographic, epidemiological and clinical variables, along with the results of laboratory analyses and imaging studies. Additionally, samples from the general population (untested for COVID-19) were also included in some analyses, with a total of 3,437 samples from the national DNA Bank and 2,506 control samples from the Genome Research at Fundación ACE (GR@ACE-Consortium). General population controls were obtained through the Spanish national blood service (Servicio Nacional de Sangre), which provides blood from unrelated adult subjects that had reported to have a Spanish origin and confirmed the absence of any personal or familial history of disease, including infectious diseases, cancer, hematologic, circulatory or endocrine diseases; mental or behavioral illnesses; or visual, hearing, respiratory, immunologic, bone, congenital, skin or digestive diseases.
Genotyping was performed on an Axiom Spain Biobank array (Thermo Fisher Scientific) in the Santiago de Compostela node of the National Genotyping Center, CeGeN (CeGen-ISCIII; http://www.usc.es/cegen). This array includes 757,836 markers and is enriched for rare variations, present in the Spanish population. Genetic data was subject to thorough quality control [excluding markers with a genotyping rate <98%, a difference in genotyping rate between cases and controls >0.02, or markers deviating from the Hardy-weinberg equilibrium. Other samples excluded from marker analysis included those with a low genotyping rate (<98%); a significantly high or low heterozygosity (deviated more than 5 deviations from the average heterozygosity); samples of related subjects and from subjects found to be non-European on an ancestry analysis (<80% European ancestry, calculated using Admixutre software)]. Additional clinical and genetic details are available in the Consortium general report, which provides complete Genome-Wide Association Study (GWAS) results for the European population (https://www.medrxiv.org/content/10.1101/2021.11.24.21266741v1). After quality control was performed, we included data for 15,045 patients of European ancestry and 588,117 genetic markers. The study was approved by regional ethics committee [Ethics Committee for Research with Medicines of Galicia, CEIm-G (No. 2020/197)] and individual consent for this retrospective analysis was waived. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
A logistic regression analysis was performed, taking case/control status as the dependent variable and being a homozygous/heterozygous carrier of the S/Z allele as the main independent variable. Disease severity was assessed in different case series and controls based on an established method.
The association study was carried out based on infection susceptibility and three severity outcomes (hospitalization, severe illness, and critical illness), which were tested using two different control definitions: A2 analysis, control samples from the general population and C analysis, COVID-19 positive individuals not meeting the case condition. Statistical analyses were performed using R software package (v4.1.2).
Results
A total of 15,045 participants were included, of whom 9,371 were COVID-19 cases and 5,674 controls (Figure 1). Median age was 62 (IQR, 50–77) and 51 (IQR, 44–61) years, respectively, and women accounted for 53.7% of patients (46.4% in controls). A1AT genotype distribution was negligible in the case of MZ and SZ, and 143 patients were homozygous SS (1%). The adjusted risk of suffering COVID-19 for homozygote SS was 1.05 (95% CI: 0.74–1.51). The adjusted risk of hospitalization for COVID-19 patients who were homozygous SS was 1.21 (95% CI: 0.77–1.88), as compared to controls, and 1.18 (95% CI: 0.71–1.98) as compared to the remainder of COVID-19 cases.
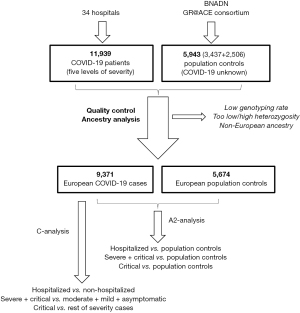
Table 1 details the clinical and demographic characteristics of European patients included in the SCOURGE study. Table 2 shows data on disease severity according to the different genotypes. The adjusted risk for severe/critical COVID-19 in individuals who are homozygous for the SS allele was 1.07 (95% CI: 0.63–1.80) as compared to controls, and 0.97 (95% CI: 0.60–1.56) as compared to the remainder of COVID-19 cases (asymptomatic, mild or moderate). As shown in Table 2, all models confirmed that patients with the PI*SS genotype do not have a higher risk of hospitalization.
Table 1
| Variable | Global (N=9,371) |
|---|---|
| Age, years, mean (SD) | 62.6 (17.9) |
| Severity, n (%) | |
| 0: asymptomatic | 582 (6.6) |
| 1: mild | 2,689 (30.3) |
| 2: intermediate | 2,099 (23.6) |
| 3: severe | 2,379 (26.8) |
| 4: critical illness | 1,128 (12.7) |
| Hospitalization, n (%) | 5,966 (63.8) |
| Severe COVID-19, n (%) | 3,507 (39.2) |
| Critical illness, n (%) | 1,128 (12.6) |
| Comorbidities, n (%) | |
| Vascular/endocrinological | 4,099 (43.7) |
| Cardiac | 1,057 (11.3) |
| Nervous | 773 (8.3) |
| Digestive | 264 (2.8) |
| Onco-hematological | 647 (6.9) |
| Respiratory | 905 (9.7) |
Table 2
| Alpha-1-antitrypsin | Cases, n (%)1 | Controls, n (%)1 | OR2 (95% CI) | OR3 (95% CI) |
|---|---|---|---|---|
| Infection risk1 | ||||
| MM | 7,531 (81.1) | 4,389 (80.8) | 1 (–) | 1 (–) |
| MS | 1,661 (17.9) | 988 (18.2) | 0.99 (0.91–1.08) | 0.94 (0.86–1.03) |
| MZ | 4 (0) | 4 (0.1) | 0.6 (0.14–2.55) | 0.7 (0.16–3.06) |
| SZ | 2 (0) | 0 (0) | – | – |
| SS | 91 (1) | 51 (0.9) | 1.06 (0.75–1.5) | 1.05 (0.74–1.51) |
| Hospitalization-related factors4 | ||||
| Hospitalization risk5, A2 analysis | ||||
| MM | 4,775 (80.5) | 4,389 (80.8) | 1 (–) | 1 (–) |
| MS | 1,094 (18.5) | 988 (18.2) | 1.03 (0.94–1.13) | 0.96 (0.86–1.08) |
| MZ | 2 (0) | 4 (0.1) | 0.48 (0.07–2.44) | 0.4 (0.05–2.45) |
| SZ | 1 (0) | 0 (0) | – | – |
| SS | 56 (0.9) | 51 (0.9) | 1.03 (0.7–1.5) | 1.21 (0.77–1.88) |
| Hospitalization risk5, C analysis | ||||
| MM | 4,775 (80.5) | 2,749 (82.1) | 1 (–) | 1 (–) |
| MS | 1,094 (18.5) | 562 (16.8) | 1.12 (1–1.26) | 1.08 (0.95–1.24) |
| MZ | 2 (0) | 2 (0.1) | 0.58 (0.07–4.81) | 0.35 (0.03–5.86) |
| SZ | 1 (0) | 1 (0) | 0.58 (0.02–14.59) | 1.15 (0.03–41.94) |
| SS | 56 (0.9) | 35 (1) | 0.92 (0.61–1.42) | 1.18 (0.71–1.98) |
| Severity risk6, A2 analysis | ||||
| MM | 2,808 (80.3) | 4,389 (80.8) | 1 (–) | 1 (–) |
| MS | 656 (18.8) | 988 (18.2) | 1.05 (0.94–1.17) | 0.99 (0.88–1.13) |
| MZ | 2 (0.1) | 4 (0.1) | 0.81 (0.11–4.17) | 0.65 (0.08–4.04) |
| SZ | 1 (0) | 0 (0) | – | – |
| SS | 31 (0.9) | 51 (0.9) | 0.97 (0.61–1.51) | 1.07 (0.63–1.8) |
| Severity risk6, C analysis | ||||
| MM | 2,808 (80.3) | 4,380 (81.4) | 1 (–) | 1 (–) |
| MS | 656 (18.8) | 944 (17.5) | 1.09 (0.97–1.21) | 1.03 (0.91–1.16) |
| MZ | 2 (0.1) | 1 (0) | 3.15 (0.3–67.72) | 4.27 (0.27–126.95) |
| SZ | 1 (0) | 1 (0) | 1.57 (0.06–39.79) | 2.72 (0.09–78.76) |
| SS | 31 (0.9) | 57 (1.1) | 0.86 (0.54–1.32) | 0.97 (0.6–1.56) |
| Critical illness risk7, A2 analysis | ||||
| MM | 897 (79.9) | 4,389 (80.8) | 1 (–) | 1 (–) |
| MS | 215 (19.1) | 988 (18.2) | 1.08 (0.91–1.27) | 1.1 (0.92–1.32) |
| MZ | 1 (0.1) | 4 (0.1) | 1.27 (0.06–8.6) | 1.11 (0.05–8.64) |
| SZ | 1 (0.1) | 0 (0) | – | – |
| SS | 9 (0.8) | 51 (0.9) | 0.88 (0.4–1.7) | 0.79 (0.34–1.66) |
| Critical illness risk7, C analysis | ||||
| MM | 897 (79.9) | 6,291 (81.1) | 1 (–) | 1 (–) |
| MS | 215 (19.1) | 1,385 (17.9) | 1.09 (0.93–1.28) | 1.07 (0.91–1.26) |
| MZ | 1 (0.1) | 2 (0) | 3.52 (0.16–36.75) | 2.95 (0.13–35.97) |
| SZ | 1 (0.1) | 1 (0) | 7.03 (0.28–177.96) | 8.88 (0.31–250.94) |
| SS | 9 (0.8) | 79 (1.0) | 0.8 (0.37–1.52) | 0.9 (0.41–1.74) |
1, values correspond to data employed in the adjusted model. 2, unadjusted logistic regression model. 3, logistic regression model adjusted by age and sex. 4, cases: COVID-19 positive; Controls: samples from the general population. 5, two different definitions of “controls” were employed: A2 analysis: control samples from the general population; and, C analysis, COVID-19 positive not satisfying the case condition. 6, cases: COVID-19 positive and hospitalized. 7, cases: COVID-19 positive with severity or critical illness. 8, cases: COVID-19 positive with critical illness. MM, homozygous allele MM; MS, heterozygous allele MS; MZ, heterozygous allele MZ; SZ, heterozygous allele SZ; SS, homozygous allele SS.
Discussion
The results of the study suggest that A1AT deficient patients are not at a higher risk for infection, severity or death from SARS-COV-2 compared to the general population.
A1AT is a serine protease inhibitor that neutralizes the proteolytic activity of elastase released by neutrophils into the lungs. A1AT deficiency increases the risk of developing conditions such as pulmonary emphysema, depending on the combination of deficient or null homozygous or heterozygous alleles encoding plasma A1AT values <60% (12). TMPRSS2 is a transmembrane protease serine that cleaves the SARS-CoV-2 spike protein to enable viral entry into cells (9). There is evidence that A1AT inhibits TMPRSS2 and that COVID-19 severity is associated with A1AT levels (13). Confirmation of this hypothesis would open an avenue for the use of serpin-based-therapies to limit COVID-19 progression and prevent SARS-CoV-2 entry into the host cell (14,15). In a recent trial, 36 patients with moderate/severe respiratory distress syndrome secondary to SARS-CoV-2 infection were randomized to receive either i.v. A1AT replacement therapy or placebo. The active treatment was biochemically effective, since it reduced serum interleukin-6 concentrations one week after administration, with good safety and patient tolerance. However, although the duration of ventilation tended to decrease in patients treated with AAT, no significant differences were observed, and mortality did not decrease (16). Therefore, exploring the potential association between A1AT deficiency and a higher risk of SARS-COV-2 infection and development of severe disease is relevant (17). A study conducted in Portugal in cohorts of patients with A1AT deficiency revealed that the Pi*ZZ genotype was associated with a significantly higher incidence of COVID-19 (7). In the same line, an Italian study demonstrated a higher frequency of SARS-COV-2 infection in the cohort of patients with severe A1AT deficiency (PI*ZZ), as compared to national infection data (8). These results suggest that this population is at a higher risk of infection and requires close monitoring.
In contrast, our case-control-study involving patients with COVID-19 and controls uncovered that A1AT deficient (PI*MS and PI*SS, prevailingly) patients and controls shared a similar risk for SARS-CoV-2 infection. These results are consistent with UKBIOBANK data (18). Although both, the UKBIOBANK study and our study were based on large population samples, no cases of PI*ZZ subjects with COVID-19 were detected in our series. Moreover, the presence of the Z allele was negligible, which limits the power of the study to detect associations. In this study, the minor allele frequency (MAF) of the Z allele was 0.0003, which is substantially below the MAF reported for the European population in the databases of reference (0.003 in the European population 1000G) (https://www.ncbi.nlm.nih.gov/snp/rs28929470). At the beginning of this century, a group of Spanish researchers explored the epidemiology of A1AT deficiency in southern Europe (France, Italy, Portugal and Spain). The authors acknowledged multiple limitations, such as very significant differences in the prevalence of carrier genes, the combination of A1AT deficiency PiS and PiZ alleles, or lack of genetic and epidemiologic data from numerous regions of the countries included in the study. All things considered, the authors estimated a prevalence of 1/496 (range, 401–6,120) for the PI*SZ genotype and 1/11,680 (range, 8,386–16,295) for the PI*ZZ genotype (19). Therefore, for these genotypes, the results obtained in our study are within the established range, since the number of cases was 9,289. In contrast, differences were observed in the PI*MZ genotype, with a lower prevalence in our series. The causes of these differences are unknown to us but, given the limitations acknowledged by the authors that hindered a reliable estimation, we cannot take their data as a valid reference to exclude results that are inconsistent with the values provided in that study. A plausible explanation is the influence of the population of Latin American ancestry living in Spain, where the frequency of the Z allele is considerably lower or even absent. In the ancestry analysis performed for data quality control, a European origin was defined as an ancestry above 80%, which allowed the inclusion of subjects of mixed origin. Nevertheless, considering the expected MAF in the European population, the power of the study is probably inadequate to detect the effect of the Z allele. A potential limitation of this study is that screening for COVID-19 was not performed in the control population in some of the assays. The use of COVID-19 controls is challenging, due to high exposure of the population to the SARS-CoV-2 virus, and the lack of data on COVID-19 status in these samples. In addition, controls may have been chosen for other purposes, and their demographic characteristics may not match with those of cases (age/sex distribution, accurate geographical location). Conversely, an advantage of using population control cohorts is that data are usually available for large samples, which increases the power of association tests (20). In addition, the GWAS performed in the SCOURGE project (11), which used the same samples as our study, demonstrated that association tests were consistent, regardless of the control population used (general population controls, controls with COVID-19 who were not hospitalized or with lower disease severity, or a combination of the two).
In summary, the population with A1AT deficiency is not at a higher risk for SARS-COV-2 infection than the general population.
Acknowledgments
Funding: This study was funded by Instituto de Salud Carlos III (COV20_00622) and co-funded by the European Union (ERDF) “A way of making Europe” program, Fundación Amancio Ortega, Banco de Santander.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1062/coif). ÁC reports funding from Instituto de Salud Carlos III-FIS-Covid and Fundación Amancio Ortega; a total of 5,000 €/year from many different institutions and entities as speaker in congresses; serving on different advisory boards from non-profit organizations; serving as president of the international Academy of Legal Medicine, and president of Kaertor Foundation. PL reports funding from Instituto de Salud Carlos III and Fundación Amancio Ortega. ARR reports funding from Instituto de Salud Carlos III. The other authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565-574. [Crossref] [PubMed]
- WHO Director-General’s opening remarks at the media briefing on COVID-19: 11 March 2020. Published March 11, 2020. Accessed 30 Mar 2020. Available online: https://www.who.int/dg/speec hes/detail/who-director-general-s-opening-remarks-at-themedia-briefingon-Covid-19---11-march-2020
- Gude-Sampedro F, Fernández-Merino C, Ferreiro L, et al. Development and validation of a prognostic model based on comorbidities to predict COVID-19 severity: a population-based study. Int J Epidemiol 2021;50:64-74. [Crossref] [PubMed]
- Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med 2020;382:1708-20. [Crossref] [PubMed]
- Maier BF, Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science 2020;368:742-6. [Crossref] [PubMed]
- Faria N, Inês Costa M, Gomes J, et al. Alpha-1 antitrypsin deficiency severity and the risk of COVID-19: A Portuguese cohort. Respir Med 2021;181:106387. [Crossref] [PubMed]
- Ferrarotti I, Ottaviani S, Balderacchi AM, et al. COVID-19 infection in severe Alpha 1-antitrypsin deficiency: Looking for a rationale. Respir Med 2021;183:106440. [Crossref] [PubMed]
- Wettstein L, Weil T, Conzelmann C, et al. Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat Commun 2021;12:1726. [Crossref] [PubMed]
- Available online: https://www.scourge-covid.org
- Cruz R, Diz-de Almeida S, López de Heredia M, et al. Novel genes and sex differences in COVID-19 severity. Hum Mol Genet 2022;31:3789-806. [Crossref] [PubMed]
- Blanco I, de Serres FJ, Fernandez-Bustillo E, et al. Estimated numbers and prevalence of PI*S and PI*Z alleles of alpha1-antitrypsin deficiency in European countries. Eur Respir J 2006;27:77-84. [Crossref] [PubMed]
- Azouz NP, Klingler AM, Callahan V, et al. Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2. Pathog Immun 2021;6:55-74. [Crossref] [PubMed]
- Dutta AK, Goswami K. Host genomics of COVID-19: Evidence point towards Alpha 1 antitrypsin deficiency as a putative risk factor for higher mortality rate. Med Hypotheses 2021;147:110485. [Crossref] [PubMed]
- Bai X, Hippensteel J, Leavitt A, et al. Hypothesis: Alpha-1-antitrypsin is a promising treatment option for COVID-19. Med Hypotheses 2021;146:110394. [Crossref] [PubMed]
- McElvaney OJ, McEvoy NL, Boland F, et al. A randomized, double-blind, placebo-controlled trial of intravenous alpha-1 antitrypsin for ARDS secondary to COVID-19. Med (N Y) 2022;3:233-248.e6. [Crossref] [PubMed]
- Yang C, Chapman KR, Wong A, et al. α1-Antitrypsin deficiency and the risk of COVID-19: an urgent call to action. Lancet Respir Med 2021;9:337-9. [Crossref] [PubMed]
- Schneider CV, Strnad P. SARS-CoV-2 infection in alpha1-antitrypsin deficiency. Respir Med 2021;184:106466. [Crossref] [PubMed]
- de Serres FJ, Blanco I, Fernández-Bustillo E. Genetic epidemiology of alpha-1 antitrypsin deficiency in southern Europe: France, Italy, Portugal and Spain. Clin Genet 2003;63:490-509. [Crossref] [PubMed]
- Mapping the human genetic architecture of COVID-19. Nature 2021;600:472-7. [Crossref] [PubMed]


