A predictive model for the identification of the risk of sepsis in patients with Gram-positive bacteria in the intensive care unit
Highlight box
Key findings
• A novel nomogram has been developed and validated to predict sepsis risk in patients with common Gram-positive bacteria, using data from the Multiparameter Intelligent Monitoring in Intensive Care IV database.
What is known and what is new?
• Numerous previous studies have been devoted to discovering potential predicted risk modes or biomarkers to detect early sepsis.
• The primary objective of this research was to create and verify a nomogram as a simple predictive model for predicting the risks of sepsis in patients with common Gram-positive bacterial infections.
What is the implication, and what should change now?
• This model performs well and might be employed clinically in the management of patients with Gram-positive bacteria.
Introduction
Sepsis is a clinical disease defined by life-threatening organ failure caused by an aberrant host response to infection, and both sepsis and septic shock are serious healthcare issues (1). Although the “Surviving Sepsis Campaign” has existed for more than 20 years, sepsis mortality and morbidity remain unacceptably high, affecting millions of people worldwide each year, with over 30% dying within 90 days (2,3). Early detection and treatment of sepsis in the initial hours following its onset can significantly reduce mortality (1). Hence, detecting early sepsis is crucial for preventing the progression from early sepsis to septic shock. However, unlike other medical emergencies, sepsis is a complex and dynamic syndrome that makes precise clinical decision-making difficult (4).
Numerous previous studies have been devoted to discovering potential predicted risk modes or biomarkers to detect early sepsis. The results of some studies have suggested that the NeoSeD score (5), and Presepsin (P-SEP) (6) may have a good performance to detect sepsis in infants. However, owing to the peculiarity of the research sample, these findings cannot be applied to a wide range of people. Some other studies have found some novel markers or methods, such as chemokine ligand 7 (CXCL7) (7), and deep learning-based model using electrocardiography (8), that may be useful in detecting early sepsis. These novel discoveries, unfortunately, cannot be widely applied due to limitations in local laboratory conditions or equipment. In addition, other research has also discovered that some factors may have a certain relationship with sepsis, but the value of these factors has not been evaluated, making it impossible to apply them appropriately in clinical decision-making (9,10).
Systemic inflammatory response syndrome (SIRS) was the first sepsis-diagnosed criteria, and it has been used as a diagnostic standard for sepsis for more than 20 years. However, with a better understanding of infectious diseases, it was revealed that some “sepsis” detected by SIRS was just an appropriate host response to infectious diseases. As a result, SIRS was abandoned due to a lack of discriminant and convergent validity (11,12). A recent case-control study discovered that the current screening score in the emergency department, known as quick Sequential Organ Failure Assessment (qSOFA), may be associated with higher mortality than SIRS. Therefore, a novel screening method for the early detection of sepsis is required (13).
The severity of sepsis is determined, in part, by the pathogen responsible for the initial infection. Initially, it was thought that the pathogens causing sepsis were predominantly Gram-negative pathogens, but more recent epidemiologic a study has shown a greater preponderance of Gram-positive pathogens (14). A study based on 10 million cases of sepsis showed that Gram-positive bacteria accounted for 52.1% of reported sepsis cases in 2000, Gram-negative bacteria accounted for 37.6%, polymicrobial infections accounted for 4.7%, anaerobes accounted for 1.0%, and fungi accounted for 4.6% (14). Meanwhile, Gram-positive, and Gram-negative bacteria are highly heterogeneous in terms of susceptible populations, susceptible sites, and mechanisms of infection (15-18). However, no studies have been conducted to predict the risk of sepsis in populations infected with Gram-positive bacteria.
Therefore, the primary objective of this research was to create and verify a nomogram as a simple predictive model for predicting the risks of sepsis in patients with common Gram-positive bacterial infections and stratifying patients into low or high-risk sepsis groups. Since Staphylococcus spp., Streptococcus spp., and Enterococcus spp. are the most prevalent gram-positive bacteria among sepsis-causing organisms, we chose infected individuals with these bacteria as our study population (19). We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1133/rc).
Methods
Sources of data
The Multiparameter Intelligent Monitoring in Intensive Care Database IV (MIMIC IV) is a large critical care database which was used to collect data for the present study. MIMIC IV is a widely available public database that contains comprehensive and high-quality data for every patient admitted to a tertiary academic medical facility in Boston, MA, USA, between 2008 and 2019 (20). This database was verified by the Institutional Review Board of the Massachusetts Institute of Technology. We were granted approval to collect data from MIMIC IV after taking the National Institutes of Health (NIH) web-based training course and the Protecting Human Research Participants examination (ID: 11218931). The MIMIC IV program was approved by the Institutional Review Boards of the Massachusetts Institute of Technology and Beth Israel Deaconess Medical Center. Patient information is anonymized so that informed patient consent is not required. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Population selection criteria
Patients infected with more than one of the three common Gram-positive bacteria on admission were considered eligible for the study. Gram-positive bacterial infections were determined by blood culture results. The three common Gram-positive bacteria included Streptococcus spp., Staphylococcus spp., and Enterococcus spp. Sepsis is characterized as severe organ failure induced by an abnormal host response to infection (11). Organ dysfunction is defined as a sudden increase in the overall SOFA score of more than 2 points as a result of the infection.
Sepsis is characterized as severe organ failure induced by an abnormal host response to infection. In this study, sepsis was defined according to Sepsis 3.0: patients with proven or suspected infection with an elevation of ≥2 SOFA points (11). Confirmed or suspected infection was determined based on ICD codes and microbiologic culture results.
The exclusion criteria were as follows: If a patient was admitted more than once, only the first stay with an available admission time was analyzed. Patients who were discharged or died within 24 hours after admission were excluded. Patients diagnosed with sepsis before or within 24 hours of admission were excluded. In the present study, patients with missing values were excluded.
Data collection and definitions
Age, gender, ethnicity, insurance, marital status, admission time, first time of being diagnosed with sepsis, discharge time, comorbidities, Glasgow coma scale (GCS), central venous pressure (CVP), SOFA score, vital signs, use of vasopressors, laboratory test results, and mechanical ventilation were all collected from MIMIC IV using Structured Query Language (SQL) in PostgreSQL (version 14.2: https://www.postgresql.org/). For multiple measurements, we used data within 24 hours of admission for analysis. If an indicator was measured multiple times within 24 hours of admission, the first measurement was used. Treatments initiated after the onset of sepsis (e.g., medications, mechanical ventilation, and dialysis) were considered ineffective.
Endpoint and follow-up time
If a patient had sepsis more than once, only time of the first diagnosis was used as the endpoint, and the period between the time of admission and the endpoint was used as the follow-up time (days). If a patient did not manifest sepsis, the discharge time was analyzed as the endpoint, and the time between the discharge time and the endpoint was the follow-up time.
Statistical analysis
Continuous and categorical variable data were reported in the form of a median with an interquartile range and a frequency with a percentage, respectively. The whole study cohort was randomly stratified into training cohorts (n=13,973) and validation cohorts (n=5,988) in a 7:3 ratio. The training cohort is used for feature selection and training of the model and the validation cohort is used to validate the performance of the model. In the training cohort, Cox regression models were used to select variables that had a significant association with the risk of sepsis. A multivariate stepwise Cox regression incorporated variables that were statistically significant in univariate Cox regression. Multivariate stepwise Cox regression is a popular method for detecting factors that are relevant to the prognosis of patients (21,22). Statistically significant variables in multivariate stepwise Cox regression were incorporated into the final multivariate Cox proportional hazards model, and the corresponding nomogram was plotted. Finally, the performance of the model was validated using receiver operating characteristic (ROC) curves, time-dependent ROC curves, calibration curves, and clinical decision curves.
R 4.0.5 software (R Foundation for statistical Computing, Vienna, Austria) was used to conduct all statistical analyses. The nomogram was depicted utilizing the “rms” package. The decision curves and calibration curves were depicted utilizing the “dcurves” package. The “survivalROC” and “riskregression” packages was used to plot ROC curves. P values less than 0.05 were deemed statistically significant.
Results
Baseline characteristics
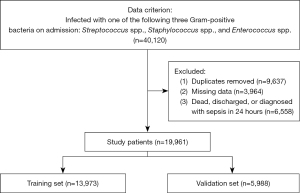
A total of 42,120 participants were admitted with three common Gram-positive bacterial infections throughout the research period. After elimination based on the exclusion criteria, 19,961 eligible patients were finally recruited (Figure 1).
Table 1 shows the details of the baseline characteristics of those patients enrolled, and a detailed comparison of the demographics and predictive variables between the training and validation cohorts. A total of 7,244 patients were infected with Streptococcus spp., 5,982 with Staphylococcus spp., and 6,735 with Enterococcus spp. A total of 19,032 patients were diagnosed with sepsis, whereas 929 patients were not diagnosed with sepsis before discharge. No significant differences were detected between the training and validation cohorts in the distribution of three Gram-positive bacterial infections, the number of sepsis, and the time of occurrence of sepsis after admission. Most patients in the overall cohort were White (66.7%). The most common coexisting disease was hypertension [13,233 (66.3%)], followed by uremia [8,534 (42.8%)], and diabetes [6,810 (34.1%)], whereas dementia, atrial fibrillation (AF), obesity, asthma, myocardial infarction, cirrhosis, burn, and chronic obstructive pulmonary disease (COPD) were present in 28.0%, 21.9%, 18.7%, 14.8%, 12.7%, 7.8%, 6.7%, and 6.6% of total patients, respectively. Tuberculous (3.1%), paralysis (1.6%), human immunodeficiency virus (HIV)/aids (1.4%), stroke (0.8%), and connective tissue disorders (0.2%) were less common. Most patients used more than 3 antibiotics [14,249 (71.4%)], whereas antifungal drug, vasopressors, and dialysis were used in 12.9%, 17.7%, and 4.8% of total patients, respectively.
Table 1
| Characteristics | Total patients (n=19,961) | Validation cohort (n=5,988) | Training cohort (n=13,973) | P |
|---|---|---|---|---|
| Gram-positive bacteria | ||||
| Streptococcus spp. | 7,244 (36.3) | 5,103 (36.5) | 2,141 (35.8) | 0.31 |
| Enterococcus spp. | 6,735 (33.7) | 4,664 (33.4) | 2,071 (34.6) | 0.102 |
| Staphylococcus spp. | 5,982 (30.0) | 4,206 (30.1) | 1,776 (29.7) | 0.544 |
| Sepsis | 929 (4.7) | 644 (4.6) | 285 (4.8) | 0.67 |
| Time (days)† | 3.88 [2.28, 6.93] | 3.88 [2.27, 6.95] | 3.88 [2.30, 6.90] | 0.917 |
| Demographics | ||||
| Gender (male) | 8,868 (44.4) | 6,211 (44.5) | 2,657 (44.4) | 0.931 |
| Age (years) | 60.00 [44.00, 73.00] | 60.00 [44.00, 73.00] | 60.00 [44.00, 73.00] | 0.691 |
| Ethnicity | 0.835 | |||
| Asian | 710 (3.6) | 502 (3.6) | 208 (3.5) | |
| Black | 3,186 (16.0) | 2,229 (16.0) | 957 (16.0) | |
| Other | 2,745 (13.8) | 1,939 (13.9) | 806 (13.5) | |
| White | 13,320 (66.7) | 9,303 (66.6) | 4,017 (67.1) | |
| Insurance | 0.722 | |||
| Medicaid | 1,849 (9.3) | 1,281 (9.2) | 568 (9.5) | |
| Medicare | 7,772 (38.9) | 5,458 (39.1) | 2,314 (38.6) | |
| Others | 10,340 (51.8) | 7,234 (51.8) | 3,106 (51.9) | |
| Complications | ||||
| Cancer | 4,386 (22.0) | 3,056 (21.9) | 1,330 (22.2) | 0.608 |
| Asthma | 2,961 (14.8) | 2,038 (14.6) | 923 (15.4) | 0.137 |
| AF | 4,372 (21.9) | 3,084 (22.1) | 1,288 (21.5) | 0.39 |
| Cirrhosis | 1,561 (7.8) | 1,091 (7.8) | 470 (7.8) | 0.944 |
| Connective tissue | 48 (0.2) | 42 (0.3) | 6 (0.1) | 0.013 |
| COPD | 1,308 (6.6) | 904 (6.5) | 404 (6.7) | 0.488 |
| Dementia | 5,583 (28.0) | 3,902 (27.9) | 1,681 (28.1) | 0.845 |
| Diabetes | 6,810 (34.1) | 4,728 (33.8) | 2,082 (34.8) | 0.209 |
| Hypertension | 13,233 (66.3) | 9,242 (66.1) | 3,991 (66.6) | 0.497 |
| Myocardial infarction | 2,544 (12.7) | 1,774 (12.7) | 770 (12.9) | 0.769 |
| Obesity | 3,724 (18.7) | 2,624 (18.8) | 1,100 (18.4) | 0.509 |
| Paralysis | 329 (1.6) | 225 (1.6) | 104 (1.7) | 0.56 |
| Renal failure | 19 (0.1) | 11 (0.1) | 8 (0.1) | 0.367 |
| Stroke | 163 (0.8) | 118 (0.8) | 45 (0.8) | 0.56 |
| Tuberculous | 622 (3.1) | 442 (3.2) | 180 (3.0) | 0.588 |
| Uremia | 8,534 (42.8) | 5,915 (42.3) | 2,619 (43.7) | 0.068 |
| Burn | 1,332 (6.7) | 915 (6.5) | 417 (7.0) | 0.295 |
| HIV | 277 (1.4) | 214 (1.5) | 63 (1.1) | 0.01 |
| Treatment | ||||
| Dialysis | 951 (4.8) | 652 (4.7) | 299 (5.0) | 0.338 |
| Antibiotics ≥3 types‡ | 14,249 (71.4) | 9,939 (71.1) | 4,310 (72.0) | 0.231 |
| Vasopressors | 3,536 (17.7) | 2,455 (17.6) | 1,081 (18.1) | 0.424 |
| Antifungal drug | 2,570 (12.9) | 1,766 (12.6) | 804 (13.4) | 0.133 |
| Antibiotic sensitivity | 0.083 | |||
| Intermediate | 343 (1.7) | 247 (1.8) | 96 (1.6) | |
| Resistant | 10,549 (52.8) | 7,340 (52.5) | 3,209 (53.6) | |
| Sensitive | 2,885 (14.5) | 1,989 (14.2) | 896 (15.0) | |
| Unknown | 6,184 (31.0) | 4,397 (31.5) | 1,787 (29.8) | |
| Ventilation | 0.226 | |||
| HFNC | 84 (0.4) | 62 (0.4) | 22 (0.4) | |
| Invasive | 3,401 (17.0) | 2,391 (17.1) | 1,010 (16.9) | |
| None | 13,430 (67.3) | 9,393 (67.2) | 4,037 (67.4) | |
| Noninvasive | 155 (0.8) | 122 (0.9) | 33 (0.6) | |
| Supplemental oxygen | 2,544 (12.7) | 1,764 (12.6) | 780 (13.0) | |
| Tracheostomy | 347 (1.7) | 241 (1.7) | 106 (1.8) | |
| Laboratory data | ||||
| Neutrophils ≥70% | 10,661 (53.4) | 7,419 (53.1) | 3,242 (54.1) | 0.179 |
| Platelet count | 0.251 | |||
| <100×109/L | 858 (4.3) | 598 (4.3) | 260 (4.3) | |
| >300×109/L | 5,763 (28.9) | 3,987 (28.5) | 1,776 (29.7) | |
| (100–300)×109/L | 13,340 (66.8) | 9,388 (67.2) | 3,952 (66.0) | |
| Creatinine | 0.828 | |||
| <0.5 mg/dL | 336 (1.7) | 233 (1.7) | 103 (1.7) | |
| >1.2 mg/dL | 4,077 (20.4) | 2,840 (20.3) | 1,237 (20.7) | |
| 0.5–1.2 mg/dL | 15,548 (77.9) | 10,900 (78.0) | 4,648 (77.6) | |
| ALT ≥40 U/L | 3,992 (20.0) | 2,815 (20.1) | 1,177 (19.7) | 0.439 |
| AST ≥40 U/L | 4,514 (22.6) | 3,155 (22.6) | 1,359 (22.7) | 0.872 |
| Total bilirubin ≥1.5 mg/dL | 1,587 (8.0) | 1,138 (8.1) | 449 (7.5) | 0.129 |
| Basophils (%) | 0.40 [0.20, 0.60] | 0.40 [0.20, 0.60] | 0.40 [0.20, 0.60] | 0.526 |
| HCO3− (mmol/L) | 26.00 [23.00, 28.00] | 26.00 [23.00, 28.00] | 26.00 [23.00, 28.00] | 0.152 |
| BUN (mg/dL) | 16.00 [12.00, 23.00] | 16.00 [12.00, 23.00] | 16.00 [11.00, 23.00] | 0.954 |
| EO (%) | 1.40 [0.60, 2.60] | 1.40 [0.60, 2.60] | 1.40 [0.60, 2.60] | 0.785 |
| Glucose (mg/dL) | 106.00 [91.00, 135.00] | 106.00 [91.00, 135.00] | 106.00 [91.00, 135.00] | 0.322 |
| HCT (%) | 37.70 [34.10, 41.10] | 37.80 [34.20, 41.10] | 37.60 [34.00, 41.00] | 0.202 |
| HGB (g/dL) | 12.60 [11.30, 13.80] | 12.60 [11.30, 13.80] | 12.60 [11.30, 13.80] | 0.237 |
| MCH (%) | 30.10 [28.60, 31.60] | 30.20 [28.60, 31.60] | 30.10 [28.60, 31.50] | 0.206 |
| MCHC (%) | 33.40 [32.40, 34.30] | 33.40 [32.40, 34.30] | 33.30 [32.40, 34.30] | 0.413 |
| Monocytes (%) | 5.50 [4.10, 7.30] | 5.50 [4.10, 7.30] | 5.50 [4.10, 7.30] | 0.562 |
| Potassium (mmol/L) | 4.10 [3.80, 4.50] | 4.10 [3.80, 4.50] | 4.10 [3.80, 4.50] | 0.793 |
| RDW (%) | 13.70 [13.10, 14.90] | 13.70 [13.10, 14.90] | 13.80 [13.10, 14.80] | 0.781 |
| RBC (×1012/L) | 4.22 [3.79, 4.61] | 4.23 [3.80, 4.61] | 4.21 [3.78, 4.61] | 0.36 |
| Sodium (mmol/L) | 139.00 [137.00, 141.00] | 139.00 [137.00, 141.00] | 139.00 [136.00, 141.00] | 0.679 |
| WBC (×109/L) | 8.10 [6.30, 10.80] | 8.10 [6.30, 10.80] | 8.10 [6.30, 10.80] | 0.919 |
| SOFA | 3.00 [2.00, 5.00] | 3.00 [2.00, 5.00] | 3.00 [2.00, 4.00] | 0.288 |
†, the time of being diagnosed with sepsis or discharge; ‡, used more than 3 types of antibiotics. Data are presented as n (%) or median [IQR]. AF, atrial fibrillation; COPD, chronic obstructive pulmonary disease; HIV, human immunodeficiency virus; HFNC, high flow nasal catheter; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HCO3−, bicarbonate; BUN, blood urea nitrogen; EO, eosinophils; HCT, hematocrit; HGB, hemoglobin; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red blood cell distribution width; RBC, red blood cell; WBC, white blood cell; SOFA, Sequential Organ Failure Assessment.
Nomogram construction and validation
A total of 8 predictive variables [the use of more than 3 types of antibiotics, dementia, ethnicity, aspartate aminotransferase (AST), neutrophils, the use of antifungal drug, ventilation and need for vasopressors] were preserved in the final simplified model and utilized to build the nomogram using stepwise selection in the Cox proportional hazards regression model. Multivariable Cox proportional regression analysis in the training cohort revealed that using more than 3 types of antibiotics [adjusted hazard ratio (HR), 2.123; 95% confidence interval (CI): 1.381–3.264; P=0.0006], needing vasopressors (adjusted HR, 1.474; 95% CI: 1.226–1.772; P<0.0001), AST ≥40 U/L (adjusted HR, 1.296; 95% CI: 1.101–1.525; P=0.0018), and neutrophil ratio ≥70% findings (adjusted HR, 1.524; 95% CI: 1.282–1.811; P<0.0001), dementia (adjusted HR, 0.540; 95% CI: 0.442–0.659; P<0.0001), need for antifungal drug (adjusted HR, 0.628; 95% CI: 0.520–0.759; P<0.0001), ventilation status and ethnicity were all independently related to an elevated risk of sepsis. Table 2 shows the associated HRs of the predictive variables in the training cohort.
Table 2
| Factors | Levels | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| Demographics | ||||||
| Age | 1.011 (1.007–1.016) | <0.0001 | ||||
| Ethnicity | Asian | – | – | – | - | |
| Black | 0.643 (0.372–1.113) | 0.1147 | 0.793 (0.458–1.373) | 0.4074 | ||
| Other | 2.030 (1.244–3.313) | 0.0046 | 1.984 (1.213–3.244) | 0.0063 | ||
| White | 1.247 (0.778–1.998) | 0.3596 | 1.235 (0.770–1.981) | 0.3821 | ||
| Gender | Female | – | – | |||
| Male | 1.212 (1.038–1.416) | 0.0153 | ||||
| Insurance | Medicaid | – | – | |||
| Medicare | 1.252 (0.927–1.689) | 0.1422 | ||||
| Others | 1.200 (0.893–1.613) | 0.2256 | ||||
| Complications | ||||||
| Asthma | 0.835 (0.652–1.069) | 0.1521 | ||||
| Burn | 0.677 (0.485–0.944) | 0.0214 | ||||
| Cancer | 0.797 (0.661–0.960) | 0.0171 | ||||
| Cirrhosis | 1.142 (0.889–1.467) | 0.2976 | ||||
| Connective tissue | 1.433 (0.357–5.747) | 0.6115 | ||||
| COPD | 1.307 (0.996–1.716) | 0.0536 | ||||
| AF | 1.854 (1.579–2.178) | <0.0001 | ||||
| Dementia | 0.649 (0.532–0.790) | <0.0001 | 0.540 (0.442–0.659) | <0.0001 | ||
| Diabetes | 0.974 (0.828–1.145) | 0.7489 | ||||
| HIV | 0.365 (0.137–0.977) | 0.0448 | ||||
| Hypertension | 1.399 (1.170–1.673) | 0.0002 | ||||
| Myocardial infarction | 1.320 (1.072–1.624) | 0.0088 | ||||
| Obesity | 1.050 (0.859–1.282) | 0.6350 | ||||
| Paralysis | 0.993 (0.547–1.802) | 0.9810 | ||||
| Renal failure | 1.372 (0.193–9.758) | 0.7519 | ||||
| Stroke | 0.449 (0.112–1.798) | 0.2577 | ||||
| Uremia | 1.407 (1.200–1.650) | <0.0001 | ||||
| Tuberculous | 1.008 (0.675–1.503) | 0.9703 | ||||
| Treatment | ||||||
| Vasopressor | 6.492 (5.534–7.616) | <0.0001 | 1.474 (1.226–1.772) | <0.0001 | ||
| Ventilation status | HFNC | – | – | – | – | |
| Invasive | 1.225 (0.653–2.300) | 0.5266 | 1.159 (0.613–2.191) | 0.6502 | ||
| None | 0.052 (0.026–0.102) | <0.0001 | 0.068 (0.035–0.136) | <0.0001 | ||
| Noninvasive | 0.294 (0.092–0.940) | 0.0390 | 0.324 (0.101–1.037) | 0.0576 | ||
| Supplemental oxygen | 0.401 (0.209–0.769) | 0.0060 | 0.444 (0.230–0.854) | 0.0150 | ||
| Tracheostomy | 2.851 (1.480–5.495) | 0.0017 | 2.894 (1.493–5.610) | 0.0017 | ||
| Antibiotic sensitivity | Intermediate | – | – | |||
| Resistant | 0.953 (0.549–1.655) | 0.8636 | ||||
| Sensitive | 0.769 (0.427–1.385) | 0.3822 | ||||
| Unknown | 0.635 (0.359–1.125) | 0.1195 | ||||
| Antibiotics ≥3 types† | 6.493 (4.275–9.86) | <0.0001 | 2.123 (1.381–3.264) | 0.0006 | ||
| Antifungal drug | 1.331 (1.107–1.600) | 0.0024 | 0.628 (0.520–0.759) | <0.0001 | ||
| Dialysis | 2.908 (2.34–3.615) | <0.0001 | ||||
| Laboratory data | ||||||
| ALT ≥40 U/L | 1.294 (1.089–1.539) | 0.0035 | ||||
| AST ≥40 U/L | 1.541 (1.311–1.810) | <0.0001 | 1.296 (1.101–1.525) | 0.0018 | ||
| Basophils | 0.761 (0.610–0.950) | 0.0160 | ||||
| BUN | 1.006 (1.002–1.010) | 0.0038 | ||||
| Creatinine | <0.5 mg/dL | – | – | |||
| >1.2 mg/dL | 0.916 (0.548–1.529) | 0.7367 | ||||
| 0.5–1.2 mg/dL | 0.757 (0.459–1.246) | 0.2734 | ||||
| Eosinophils | 0.939 (0.900–0.979) | 0.0034 | ||||
| Glucose | 1.001 (1. 000–1.002) | 0.2485 | ||||
| HCO3− | 0.972 (0.953–0.992) | 0.0053 | ||||
| HCT | 1.009 (0.996–1.021) | 0.1815 | ||||
| Hemoglobin | 1.012 (0.977–1.049) | 0.4975 | ||||
| MCH | 1.016 (0.990–1.041) | 0.2282 | ||||
| MCHC | 0.947 (0.903–0.993) | 0.0239 | ||||
| Monocytes | 0.976 (0.954–0.999) | 0.0409 | ||||
| Neutrophils ≥70% | 1.878 (1.582–2.228) | <0.0001 | 1.524 (1.282–1.811) | <0.0001 | ||
| Platelet count | <100×109/L | – | – | |||
| >300×109/L | 0.762 (0.552–1.052) | 0.0991 | ||||
| (100–300)×109/L | 1.036 (0.772–1.391) | 0.8141 | ||||
| Potassium | 1.015 (0.907–1.134) | 0.7996 | ||||
| RDW | 1.000 (0.964–1.037) | 0.9983 | ||||
| RBC | 0.992 (0.893–1.102) | 0.8804 | ||||
| Sodium | 1.000 (0.981–1.019) | 0.9918 | ||||
| Total bilirubin ≥1.5 mg/dL | 1.516 (1.226–1.875) | 0.0001 | ||||
| WBC | 1.001 (0.996–1.006) | 0.6889 | ||||
| SOFA | 1.111 (1.088–1.136) | <0.0001 | ||||
†, used more than 3 types of antibiotics. HR, hazard ratio; CI, confidence interval. AF, atrial fibrillation; COPD, chronic obstructive pulmonary disease; HIV, human immunodeficiency virus; HFNC, high flow nasal catheter; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HCO3−, bicarbonate; BUN, blood urea nitrogen; HCT, hematocrit; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red blood cell distribution width; RBC, red blood cell; WBC, white blood cell; SOFA, Sequential Organ Failure Assessment.
In the total study cohort, the incidence of sepsis in patients was 4.9%, 4.5%, 4.6% and 4.7% at 3-day, 1-week, 2-week, and 1-month, respectively. The nomogram for predicting 3-day, 1-week, 2-week, and 1-month sepsis risk is depicted in Figure 2. The nomogram was created by proportionately translating each regression coefficient in multivariable Cox analysis to a 0 to 100 points scale. Each covariate among the 8 variables that comprise the nomogram was given a score by drawing a matching vertical line straight down to the axis labeled points. Individual probabilities of 3-day, 1-week, 2-week, and 1-month sepsis may be calculated by summing the entire score and determining its position on the total points scale. For each variable included in the model, we plotted Kaplan-Meier survival curves based on their grouping. The frequency of sepsis can be seen in the risk table below the curve (Figure 3).


Area under the curve (AUC), calibration plots, and decision plots were utilized to examine the performance of this nomogram. In both the training and validation cohorts, we generated ROC curves for 3-day, 1-week, 2-week, and 1-month sepsis risk, with the AUC value displayed (Figure 4). The AUC for predicting 3-day, 1-week, 2-week, and 1-month sepsis risk in the training cohort was 0.857, 0.774, 0.740, and 0.728, respectively (Figure 4A-4D), and was 0.855, 0.781, 0.742, and 0.742, respectively, in the validation cohort (Figure 4E-4H), showing that the model had strong discrimination. We also generated the ROC curves for 3-day, 1-week, 2-week, and 1-month sepsis risk for 3 Gram-positive bacterial infections in the test cohort (Figure 4I-4K). The model has good predictive performance in different Gram-positive bacteria. In the patients infected with Streptococcus spp., the model had AUCs of 0.909, 0.77, 0.689, and 0.685 at 3-day, 1-week, 2-week, and 1-month, respectively. In the patients infected with Enterococcus spp., the model had AUCs of 0.818, 0.786, 0.765, and 0.796 at 3-day, 1-week, 2-week, and 1-month, respectively. In the patients infected with Staphylococcus spp., the model had AUCs of 0.85, 0.778, 0.742, and 0.746 at 3-day, 1-week, 2-week, and 1-month, respectively. In addition, we plotted time-dependent ROC curves in the training and validation cohorts. The results show that the AUC of the model is higher than 0.8 regardless of the prediction at any time point (Figure 4L,4M). The calibration plots also revealed consistency between the predicted probability of morbidity and actual observation, indicating that the model was well-calibrated (Figure 5). Clinical decision curve analysis suggested that most patients with Gram-positive bacterial infections could benefit from the prediction model no matter whether at 3-day, 1-week, 2-week, and 1-month (Figure 6).



Discussion
A novel nomogram model for predicting 3-day, 1-week, 2-week, and 1-month probabilities of sepsis in patients admitted to the intensive care unit (ICU) and infected with three types of common Gram-positive bacterial infections (Streptococcus spp., Enterococcus spp., and Staphylococcus spp.) was developed and validated in this study using multivariate stepwise Cox regression. This study revealed three important results: (I) in multivariable analysis, the use of more than 3 types of antibiotics, dementia, ethnicity, AST, neutrophils, the use of antifungal drug, ventilation and need for vasopressors were independently related to the risk of sepsis; (II) the nomogram model demonstrated strong prediction capacity and risk stratification abilities; and (III) the nomogram model showed a lot of potential for guiding treatment decisions.
The nomogram model demonstrated strong discriminative potential for early sepsis (AUC of 0.857 and 0.855 in the development and validation cohorts on the third day, respectively). The nomogram has a simple visual interface where the “total points” is calculated based on the “points” of each variable. The “total points” vertically corresponds to the scale on the predictor below, which is the patient’s risk of developing sepsis. Thus, clinicians can easily identify high-risk patients. Additionally, the calibration plots revealed that the predicted 3-day, 1-week, 2-week, and 1-month sepsis probability closely matched the actual sepsis probabilities in both the development and validation cohorts, demonstrating the model’s calibration ability. The calibration curves of three different species of Gram-positive bacteria in the validation cohort demonstrated that the nomogram model had high consistency in the prediction performance of different Gram-positive bacteria, with no obvious overestimation or underestimation of risk. Clinical usefulness is a crucial indicator in evaluating a prediction model’s applicability and benefits to patients in clinical settings. Hence, to validate the model’s clinical use, we utilized clinical decision curves to determine if nomogram-assisted decisions would enhance patient outcomes. The results revealed that, in both the development and validation cohorts, nomogram-assisted decisions resulted in a larger net benefit as compared to treatment of all patients or none.
Having used more than 3 types of antibiotics may be associated with being critically ill. The Surviving Sepsis Campaign in 2016 recommends empiric broad-spectrum treatment with one or more antibacterial drugs for patients with sepsis or septic shock to cover all suspected infections (including bacterial and potentially fungal or viral coverage). One of the most essential reported aspects of the successful management of life-threatening infections causing sepsis and septic shock is the initiation of appropriate antimicrobial therapy (i.e., with activity against the causative pathogen or pathogens), and failure to initiate appropriate empiric therapy in patients with sepsis and septic shock is related to a significant increase in morbidity and mortality (23). Since then, based on sepsis guidelines, clinicians have frequently combined more than 2 or 3 types of antibiotics in clinical practice for suspected sepsis and septic shock. As a result, we can conclude that if patients received more than 3 types of antibiotics from their physicians, they had been considered critically ill. Conversely, combination therapy of multi-antibiotics may enhance drug toxicity, aggravate organ damage, and even promote the development of sepsis. In addition, two large observational series and a randomized controlled trial suggested that rapidity of treatment with antibiotics was the most important matter for patients who have septic shock, but whether it has benefits for patients who have sepsis without shock remained controversial (24,25). In our study, we also discovered that patients who were infected with bacteria and received more than 3 types of antibiotics were more likely to have sepsis. To make our clinical decisions more appropriate and limit the side effects of antibiotic combinations, we should obtain more information before deciding which antibiotics to use for patients with suspected sepsis but without shock, rather than choosing more than 3 types of antibiotics blindly and hurriedly.
We discovered that patients with dementia were less likely to develop sepsis in the ICU than those without dementia, which was consistent with a previous study. A large-sample study of 148,293 people found that patients with dementia had a lower risk of comorbidity burden and organ failure than those without dementia because of more frequent medical monitoring and that in cases of organ failure, the dementia cohort received less invasive treatment measures (26). However, many previous studies have indicated that organ dysfunction, comorbidity, and invasive treatment are important factors in the pathogenesis of sepsis (27-29). However, another study showed that dementia was a risk factor for in-hospital death in patients with sepsis. But the patients in this study had been diagnosed with sepsis and the outcome indicator was in-hospital death, which is different from our study design (30).
Furthermore, needing vasopressors may be associated with sepsis risk. The SOFA score comprises an evaluation of cardiovascular function (11). Patients with septic shock have chronic hypotension needing vasopressors to maintain mean arterial pressure (MAP) more than 65 mmHg and a blood lactate level greater than 2 mmol/L despite appropriate volume resuscitation (11). This is consistent with our findings.
We discovered that different races are linked with varying sepsis risks. Blacks were shown to have the lowest risk compared to Asians and Whites, which was similar to some recent studies. A large sample size retrospective analysis of 1,114,386 sepsis patients indicated that Blacks had significantly lower sepsis risk and hospital mortality than Whites (31). Another study in Michigan discovered that African Americans had the lowest hospital mortality, whereas Asian-Americans were in the middle and Whites had the highest hospital mortality among sepsis patients (32). However, a review concluded that sepsis occurrence was much higher among Black people compared to non-Hispanic White people because Black people have less formal education, no insurance, and a lower income (33). An international, multicenter study may be required to explain the differences between these studies. Since our study population was from hospitals in the United States, further external validation is needed for the applicability of our conclusions to other regions.
Neutrophils are the most abundant type of white blood cells in humans and serve as the host’s first line of defense against invading pathogens. However, we discovered that a neutrophil ratio greater than 70% was correlated with an elevated risk of sepsis in patients infected with Gram-positive bacteria. The production of proteases and reactive oxygen species by neutrophils can lead to hyperinflammation in sepsis (34). The improper activation and placement of neutrophils inside the microvasculature induce multiple organ failures (35,36). For example, Park et al. found that neutrophil aggregates could lead to the generation of dead space in the pulmonary microcirculation, then lead to acute lung injury (ALI) (37). Marki et al. also found that elongated neutrophil-derived structures were 10- to 100-fold more prevalent in the blood plasma of septic patients than in healthy donors (38). Recently, several studies have noted that an increase in plasma protease activity and protein hydrolysis coincided with recurrent bacterial infections, indicating that dysregulation of protein hydrolysis may lead to adverse outcomes (39,40). Another earlier study indicated that protein hydrolyzing enzymes stimulate neutrophil apoptosis. Combined, it seems likely that the increase in protein hydrolysis is a secondary response to the increase in neutrophils, both of which portend an increased risk of sepsis and a poor prognosis (41).
This study found several significant risk factors associated with the risk of sepsis, constructed a predictive nomogram for clinical use, and validated it by ROC curves, calibration curves, and clinic decision curves. In the present study, the included patients were not limited by age and sex, so the conclusions of this study apply to a wide range of populations. This study only included patients infected with three common Gram-positive bacteria, which restricted the types of bacteria studied, possibly reducing the impact of differences in pathogenicity among bacteria on the risk of sepsis and clarifying the appropriate population for this study. The factors included in this study were common clinical complications and commonly used laboratory indicators, which increases the clinical practicability of the model.
There were some limitations to the present study. First, there is always the possibility of bias in retrospective studies. Consequently, the nomogram was built with a high sample size cohort to decrease bias. Second, despite the use of multivariable Cox regression models, residual confounding and unquantified factor confounding cannot be excluded. Third, some indicators of interest to us, such as calcitoninogen and interleukin-6 (IL-6), were not included in the data analysis due to the fact that some of the data in the public databases were missing too much. Finally, even though internal validation demonstrated that the nomogram model had high discrimination and calibration, we did not conduct external validation. Hence, we intend to conduct external validation utilizing additional ethnic communities and places with a bigger scale sample size.
Conclusions
In summary, we managed to build a predicted model and constructed a nomogram to predict the sepsis possibility in patients with some common types of Gram-positive bacteria, especially Streptococcus spp., Enterococcus spp., and Staphylococcus spp. This model performs well and might be employed clinically in the management of patients with Gram-positive bacteria.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1133/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1133/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1133/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Evans L, Rhodes A, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Crit Care Med 2021;49:e1063-143. [Crossref] [PubMed]
- Yende S, Austin S, Rhodes A, et al. Long-term quality of life among survivors of severe sepsis: analyses of two international trials. Crit Care Med 2016;44:1461-7. [Crossref] [PubMed]
- Levy MM, Evans LE, Rhodes A. The Surviving Sepsis Campaign Bundle: 2018 update. Intensive Care Med 2018;44:925-8. [Crossref] [PubMed]
- Tulli G, Toccafondi G. Integrating infection and sepsis management through holistic early warning systems and heuristic approaches: a concept proposal. Diagnosis (Berl) 2021; Epub ahead of print. [Crossref]
- Sokou R, Ioakeimidis G, Piovani D, et al. Development and validation of a sepsis diagnostic scoring model for neonates with suspected sepsis. Front Pediatr 2022;10:1004727. [Crossref] [PubMed]
- Botondi V, D'Adamo E, Plebani M, et al. Perinatal presepsin assessment: a new sepsis diagnostic tool? Clin Chem Lab Med 2022;60:1136-44. [Crossref] [PubMed]
- Pociute A, Kottilingal Farook MF, Dagys A, et al. Platelet-Derived Biomarkers: Potential Role in Early Pediatric Serious Bacterial Infection and Sepsis Diagnostics. J Clin Med 2022;11:6475. [Crossref] [PubMed]
- Kwon JM, Lee YR, Jung MS, et al. Deep-learning model for screening sepsis using electrocardiography. Scand J Trauma Resusc Emerg Med 2021;29:145. [Crossref] [PubMed]
- Wallgren UM, Larsson E, Su A, et al. Keywords reflecting sepsis presentation based on mode of emergency department arrival: a retrospective cross-sectional study. Int J Emerg Med 2021;14:78. [Crossref] [PubMed]
- Lu G, Zhou J, Yang T, et al. Landscape of Metabolic Fingerprinting for Diagnosis and Risk Stratification of Sepsis. Front Immunol 2022;13:883628. [Crossref] [PubMed]
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- Govindan S, Iwashyna TJ. Web Exclusives. Annals for Hospitalists Inpatient Notes - Sepsis-3 for Hospitalists-Sepsis Without SIRS. Ann Intern Med 2016;165:HO2-3. [Crossref] [PubMed]
- Robert Boter N, Steinherr Zazo A, Rocamora Blanch G, et al. Sepsis Code in emergency department: Performance of q-SOFA score compared to SIRS score. Med Clin (Barc) 2022;158:260-4. [Crossref] [PubMed]
- Martin GS, Mannino DM, Eaton S, et al. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 2003;348:1546-54. [Crossref] [PubMed]
- Li C, Wang J, Wang Q, et al. Predictive Value of a Quick Pitt Bacteremia Score for Prognosis of Patients with Bloodstream Infection Secondary to Urinary Tract Infection: A Retrospective Cohort Study. Infect Drug Resist 2022;15:4381-91. [Crossref] [PubMed]
- Xu CY, Ye HW, Chen B, et al. Analysis of risk factors and prognosis of post-stroke pulmonary infection in integrated ICU. Eur Rev Med Pharmacol Sci 2021;25:856-65. [Crossref] [PubMed]
- Li B, Gao Y, Wang X, et al. Clinical features and outcomes of bacterascites in cirrhotic patients: A retrospective, multicentre study. Liver Int 2020;40:1447-56. [Crossref] [PubMed]
- Feezor RJ, Oberholzer C, Baker HV, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun 2003;71:5803-13. [Crossref] [PubMed]
- Vincent JL, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009;302:2323-9. [Crossref] [PubMed]
- Johnson A, Bulgarelli L, Pollard T, et al. MIMIC-IV. PhysioNet 2020. Available online: https://physionet.org/content/mimiciv/1.0/ (accessed August 23, 2021).
- Jin S, Xie L, You Y, et al. Development and validation of a nomogram to predict B-cell primary thyroid malignant lymphoma-specific survival: A population-based analysis. Front Endocrinol (Lausanne) 2022;13:965448. [Crossref] [PubMed]
- Powell CA, Modi S, Iwata H, et al. Pooled analysis of drug-related interstitial lung disease and/or pneumonitis in nine trastuzumab deruxtecan monotherapy studies. ESMO Open 2022;7:100554. [Crossref] [PubMed]
- Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Crit Care Med 2017;45:486-552. [Crossref] [PubMed]
- Buckman SA, Turnbull IR, Mazuski JE. Empiric Antibiotics for Sepsis. Surg Infect (Larchmt) 2018;19:147-54. [Crossref] [PubMed]
- Mi MY, Klompas M, Evans L. Early Administration of Antibiotics for Suspected Sepsis. N Engl J Med 2019;380:593-6. [Crossref] [PubMed]
- Bouza C, Martínez-Alés G, López-Cuadrado T. The impact of dementia on hospital outcomes for elderly patients with sepsis: A population-based study. PLoS One 2019;14:e0212196. [Crossref] [PubMed]
- Martin GS, Mannino DM, Moss M. The effect of age on the development and outcome of adult sepsis. Crit Care Med 2006;34:15-21. [Crossref] [PubMed]
- Lagu T, Rothberg MB, Shieh MS, et al. What is the best method for estimating the burden of severe sepsis in the United States? J Crit Care 2012;27:414.e1-9. [Crossref] [PubMed]
- Bouza C, López-Cuadrado T, Amate-Blanco JM. Characteristics, incidence and temporal trends of sepsis in elderly patients undergoing surgery. Br J Surg 2016;103:e73-82. [Crossref] [PubMed]
- Daw MA, El-Bouzedi AH, Dau AA. Trends and patterns of deaths, injuries and intentional disabilities within the Libyan armed conflict: 2012-2017. PLoS One 2019;14:e0216061. [Crossref] [PubMed]
- Chaudhary NS, Donnelly JP, Wang HE. Racial Differences in Sepsis Mortality at U.S. Academic Medical Center-Affiliated Hospitals. Crit Care Med 2018;46:878-83. [Crossref] [PubMed]
- Engoren M, Arslanian-Engoren C. Race and sex based disparities in sepsis. Heart Lung 2022;52:37-41. [Crossref] [PubMed]
- Minejima E, Wong-Beringer A. Impact of Socioeconomic Status and Race on Sepsis Epidemiology and Outcomes. J Appl Lab Med 2021;6:194-209. [Crossref] [PubMed]
- van der Poll T, Shankar-Hari M, Wiersinga WJ. The immunology of sepsis. Immunity 2021;54:2450-64. [Crossref] [PubMed]
- Brown KA, Brain SD, Pearson JD, et al. Neutrophils in development of multiple organ failure in sepsis. Lancet 2006;368:157-69. [Crossref] [PubMed]
- Hong Y, Chen L, Sun J, et al. Single-cell transcriptome profiling reveals heterogeneous neutrophils with prognostic values in sepsis. iScience 2022;25:105301. [Crossref] [PubMed]
- Park I, Kim M, Choe K, et al. Neutrophils disturb pulmonary microcirculation in sepsis-induced acute lung injury. Eur Respir J 2019;53:1800786. [Crossref] [PubMed]
- Marki A, Buscher K, Lorenzini C, et al. Elongated neutrophil-derived structures are blood-borne microparticles formed by rolling neutrophils during sepsis. J Exp Med 2021;218:e20200551. [Crossref] [PubMed]
- Maegele M, Aletti F, Efron PA, et al. New insights into the pathophysiology of trauma and hemorrhage. Shock 2023;59:6-9. [Crossref] [PubMed]
- Bauzá-Martinez J, Aletti F, Pinto BB, et al. Proteolysis in septic shock patients: plasma peptidomic patterns are associated with mortality. Br J Anaesth 2018;121:1065-74. [Crossref] [PubMed]
- Trevani AS, Andonegui G, Giordano M, et al. Neutrophil apoptosis induced by proteolytic enzymes. Lab Invest 1996;74:711-21.
(English Language Editor: J. Jones)