
Diagnostic and treatment value of XPNPEP3 in acute myocardial infarction
Highlight box
Key findings
• An XPNPEP3 gene and diagnosis model based on XPNPEP3 expression in acute myocardial infarction (AMI) were established, which may be useful for assessing curative effects in AMI.
What is known and what is new?
• AMI is a serious cardiovascular disease with high morbidity and mortality.
• XPNPEP3 gene considered for the first time to be able to serve as a diagnostic marker for AMI.
What is the implication, and what should change now?
• An XPNPEP3 gene and diagnosis model based on XPNPEP3 expression in AMI may be used for clinical application in AMI to help clinicians to develop personalized treatment.
Introduction
Acute myocardial infarction (AMI) is a multifactorial coronary artery flow blockage based on cardiac atherosclerosis and thrombosis, which leads to the interruption of blood supply to cardiomyocytes and then causes acute necrosis of local cardiomyocytes, endangering the patient’s life (1,2). As one of the most common critical cardiovascular diseases, AMI has high morbidity and mortality (3). Early diagnosis, correct evaluation, and timely reperfusion has great significance in mortality reduction as well as prognostic improvement (4,5). The clinical diagnosis of AMI is mainly based on the patient’s symptoms, vital signs, electrocardiogram, and myocardial enzyme spectrum. However, the changes of these indicators are not obvious in some patients, which may easily lead to misdiagnosis and missed diagnosis (6,7). Creatine kinase-MB isoenzyme (CK-MB) and cardiac troponin I (cTnI) have been commonly determined, but their sensitivity cannot meet clinical needs. Therefore, it is necessary to develop new diagnostic and therapeutic drugs to complement clinical applications.
MicroRNAs (miRNAs) are a class of small RNAs with a length of about 20–24 nucleotides, which play various important regulatory roles in cells. They do not have the function of protein encoding, however, they can simultaneously influence epigenetics, post-transcription, and transcription of genes, to promote messenger RNA (mRNA) degradation or inhibit protein translation (8). miRNAs are abundant and stable in the blood, and they can be aberrantly expressed under pathological conditions of cardiovascular disease (9). In addition, Chen et al. showed that miRNAs play a key role in the diagnosis of AMI and its associated symptomatic diseases, platelet activation monitoring, and prognosis prediction (9). Cardiac-rich miRNAs (such as miR-499, miR-208, and miR-1) were able to be rapidly upregulated in repertoire plasma after myocardial necrosis (10,11). However, in-depth study of the molecular mechanism of miRNA in AMI is relatively rare, and the exact molecular network underlying the regulatory mechanism mediated by miRNA during AMI progression has remained unclear.
In this study, we searched the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geoprofiles) to obtain public data sets (GSE24584, GSE24519), and explored the integration of miRNAs and mRNA expression spectrum results. Between the AMI group and the normal control group, differentially expressed mRNAs (DEmRNAs) and miRNAs (DEmiRNAs), were filtered. Subsequently, a miRNA-mRNA regulatory network was developed. The use of a protein-protein interaction (PPI) network and molecular docking helped to further identify key drugs. Comprehensive analysis applying bioinformatics studies were conducted with the aim to discover therapeutic targets for AMI and diagnostic markers. We present this article in accordance with the STREGA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1203/rc).
Methods
Raw data
The GEO database was used to determine the miRNA expression of the GSE24548 dataset (12), including 3 normal samples and 4 AMI samples, and the mRNA expression of the GSE24519 dataset (13), including 4 normal samples and 34 AMI samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DEmiRNA/DEmRNA
The limma package in R (14) was used to screen DEmiRNA in the GSE24548 dataset and DEmRNA [differentially expressed genes (DEGs)] in the GSE24519 dataset, with P<0.05 and |log2(fold change)| >1 as the threshold value.
Regulatory networks of miRNA-mRNA
The target genes (DEGs) of miRNA were obtained by TargetScan (https://www.targetscan.org/vert_80/), and relationship pairs (miRNA-mRNA) were plotted by Cytoscape (https://cytoscape.org/). Cytoscape is software that can integrate biomolecular interaction networks with high-throughput expression data and other molecular states into a unified framework that can be adapted to any system of molecular components and interactions (15).
Functional enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation was carried out in the R Package ClusterProfiler (16).
Identification of hub genes
The following analyses were conducted to identify the hub genes. ESTIMATEScore of samples were determined by Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) (17), and Hmisc package was used to calculate Spearman analysis between ESTIMATEScore and DEGs. Next, the Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) (18) algorithm evaluates gene expression profiling datasets to obtain 22 immune cell infiltration scores per patient. The Hmisc package was used to conduct Spearman analysis between immune cells score and DEGs.
Gene set variation analysis (GSVA)
In the gene set enrichment analysis (GSEA) website, KEGG pathway-related gene sets (c2.Cp.KEGG.V7.0.Symbols.GMT) were downloaded, and then the GSVA package was used to calculate score of each pathway. Next, the Hmisc package was used to screen hub gene related pathways with P<0.05 and |correlation (cor)| >0.3.
Classification algorithms
In the GSE24519 dataset, based on the identified hub genes, the RMS package in R was used to build a diagnostic model.
Screening of candidate drugs
Based on the drug target pair in drugbank and the PPI network (threshold score of 400), the proximity of the drug and treatment for AMI was calculated. Here, we can give S (that is, the gene set associated with the treatment of AMI), D (that is, the degree of the node of the gene set associated with PPI), T (the drug target gene set), and distance d(s,t) as the shortest path between s node and t node (where s∈S, the gene related to heart failure; t∈T, is the drug target gene), the calculation method is as follows:
Where ω is the weight of the target gene. If the target gene is a gene in the AMI related gene set, the calculation method is ω = −ln(D + 1), otherwise ω=0.
The simulated reference distance distribution corresponding to the drug was generated. Simply speaking, a set of protein nodes were randomly selected as the simulated drug target in the network, and the number of nodes was the same as the target size (represented by R). Then calculate the distance d(S,R) between these simulated drug targets (representing the simulated drug) and the relevant gene set of the key gene, and generate the simulated reference distribution after 10,000 random repetitions, and convert the mean and standard deviation of the μd(S,R) and σd(S,R) reference distributions and corresponding to the actual desired observation distances into standardized scores. That is, proximity z:
Regardless of whether the relevant gene sets of key genes were taken as samples or the randomly selected gene sets were taken as samples, multiple hypothesis tests were carried out based on the random data obtained in reference and drugs with small distance and false discovery rate (FDR) <0.001 were selected as drug candidates.
Molecular docking simulation
Autodock Vina software was used for molecular docking simulation (19). AutoDockTools 1.5.6 (20) was used to prepare all of the input files. From the Protein Data Bank (PDB) (21) database, the PDB-IDs of target genes were acquired. Identification of the strongest binding mode to ligand molecules was performed using Lamarckian algorithm (22), where, in molecular docking, the grid coordinates in each of the XYZ directions are −10, 18.5 and 13.5, respectively, and the grid length in each direction is 20Å. Specifically, the maximum allowable energy was 3 kcal/mol, the exhaustiveness was 8, and the maximum number of conformations output was 10. Results processing was performed in Pymol (23). Using the Gromacs2019 software package (24), we conducted 100 nanoseconds (ns) molecular dynamics simulations for assessing receptor-ligand complex for its binding stability.
Statistical analysis
The statistical data of this study were all obtained using the R software (version 3.6.0). The Sangerbox platform supported all the analyses of this study (25). Notably, P<0.05 was considered statistically significant.
Results
Identification of DEmiRNA
In the GSE24548 dataset, 34 upregulated miRNAs and 2 downregulated miRNAs were screened (Figure 1A,1B). TargetScan analysis showed that only 31 DEmiRNAs had target genes. Furthermore, KEGG analysis showed that classical pathways were enriched in 31 DEmiRNAs (Figure 1C).

Construction of a miRNA-mRNA network
In the GSE24519 dataset, 57 upregulated mRNA and 6 downregulated mRNA were screened (Figure 2A,2B). Among 63 DEGs, only 47 DEGs were identified as target genes of 31 miRNAs. Next, a miRNA-mRNA network including 31 miRNAs and 47 mRNAs was constructed by Cytoscape (Figure 2C). Network topology analysis showed the significance degree of 47 DEGs (Figure 2D).
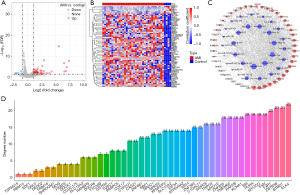
Identification of hub XPNPEP3 gene
The correlation analysis between ESTIMATEScore and 47 DEGs showed that 6 DEGs were associated with ESTIMATEScore (Figure 3A). Furthermore, the correlation analysis between 22 kinds of immune scores and 6 DEGs showed that XPNPEP3 was most closely associated with immune cells (Figure 3B).

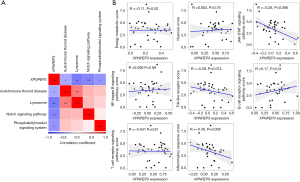
Inflammatory response pathway was associated with XPNPEP3 gene
GSVA analysis showed that 4 hub pathways (AUTOIMMUNE_THYROID_DISEASE, LYSOSOME, NOTCH_SIGNALING_PATHWAY, PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM) may potentially be regulated by XPNPEP3 (Figure 4A). At the same time, we analyzed the relationship between XPNPEP3 and the scores of pathways related to energy metabolism, hypoxia, and inflammation (Figure 4B), and we finally found that XPNPEP3 was negatively correlated with pathways related to inflammatory response.
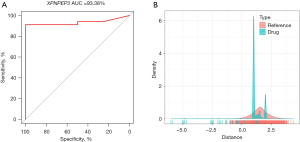
DB06909 was a potential inhibitor of XPNPEP3
The above analysis indicated that the role of the XPNPEP3 gene in AMI patients was worthy of further research. Therefore, in the GSE24519 dataset, a diagnostic model was built according to the expression of XPNPEP3 with an area under the curve (AUC) of 93.38% (Figure 5A). Correlation analysis of AMI patients in the GSE24519 dataset was conducted in the rcorr function of the Hmisc package, and a total of 159 genes were found to highly significantly correlate with the XPNPEP3 gene. The PPI analysis showed the distance density distribution of drugs to XPNPEP3-association gene sets (Figure 5B).
Molecular docking was conducted to confirm whether the 7 compounds showing the closest relation to the XPNPEP3 gene set (Table 1) also exert great regulatory effects on the XPNPEP3 protein. In the network, all the key compounds, especially DB06909, showed a strong affinity to XPNPEP3 protein (−7.6 kcal/mol) (Figure 6A). Additionally, DB06909 could generate a stable complex with XPNPEP3 via hydrogen bonding to ASP118, SER210, and ASN102 of the XPNPEP3 protein and hydrophobic interaction with ASP205, VAL99, and TYR105 (Figure 6B).
Table 1
| Compounds | Score | H-bond interactions | Hydrophobic interactions |
|---|---|---|---|
| DB06909 | −7.6 | ASP118, SER210, ASN102 | ASP205, VAL99, TYR105 |
| DB02660 | −6.8 | ASP342, ASP331, HIS424, PRO301 | VAL430, VAL303, TYR300, HIS431, HIS314 |
| DB01113 | −6.7 | HIS431, HIS314 | HIS420, GLU451, VAL430 |
| DB01647 | −6.7 | ARG438, HIS431, LEU313 | HIS314, GLU451, TYR300, VAL430 |
| DB03807 | −6.6 | – | TYR300, VAL430, VAL303, LEU313, HIS420 |
| DB02095 | −5.9 | HIS431, HIS424, ASP331 | VAL430 |
| DB02211 | −5.9 | ASP331 | TYR300, HIS431, VAL430, LEU313, HIS314 |
ASP, aspartate; SER, serine; ASN, asparagine; VAL, valine; TYR, tyrosine; HIS, histidine; PRO, proline; GLU, glutamate; ARG, arginine; LEU, leucine.
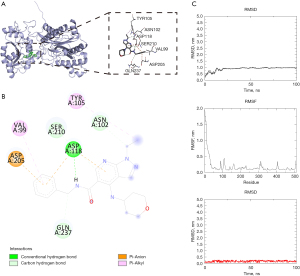
The XPNPEP3 protein concept was stable, as shown by molecular dynamics simulations at 100 ns (Figure 6C). Generally, compound DB06909 bound relatively stably to the active site of XPNPEP3 protein. This demonstrated a high potential of compound DB06909 serving as an inhibitor to XPNPEP3 protein.
Discussion
Rapid diagnosis of AMI is important for the management of patients with chest pain. There have been many studies based on a variety of technological tools and screening biomarkers to provide critical information on the accuracy of the diagnosis of AMI and its associated diseases (26-28). By using bioinformatic target prediction to integrate mRNA and miRNA expression data, it helped us to realize that upregulated miRNAs may largely contribute to the transcriptome of downregulated mRNAs (29). A comprehensive analysis provided evidence for a possible regulatory crossover between cardiac miRNAs and the Nrf2 transcriptional network (30). Wu et al. reported that a miRNA-mRNA regulatory network including 10 key genes and 3 miRNAs and 5 potential drugs that may provide treatment for chronic Chagas cardiomyopathy (31). A study screened 5 genes (POSTN, SFRP2, LOX, TIMP1, and SPARC) associated with myocardial infarction using analysis combining miRNA and mRNA microarray (32). Chen et al. showed that a competing endogenous RNA (ceRNA) regulatory network including 4 related circRNAs (cirC_0110609, cirC_0002702, cirC_0047959, cirC_0013751), 4 miRNAs (miR-378a-3p, miR-342-3p, miR-27b-3p, miR-20a-5p), and 3 mRNAs (Thbs1, Tgfbr3, Col12a1) could function critically in cardiac hypertrophy (33). Herein, we constructed a miRNA-mRNA network including 31 DEmiRNAs and 47 DEGs, and identified that the XPNPEP3 gene was most associated with AMI. The data demonstrated that XPNPEP3 may participate in AMI initiation and progression.
Furthermore, a diagnostic model based on XPNPEP3 expression showed an AUC of 93.38%, which indicated that XPNPEP3 had vital influence in AMI. XPNPEP3 encodes X-proline aminopeptidase 3, which is an enzyme that works through the removal of N-terminal proline residue from peptides. In regulating normal ciliary function, XPNPEP3 plays an important role and its deficiency could lead to nephritis-like ciliopathy (34). A study showed that ectopic expression of XPNPEP3 promoted tumorigenic properties in colorectal cancer cells (35). Based on multi-omics, another study verified that XPNPEP3 had increased expression in esophageal squamous cell carcinoma (17). At present, the role of XPNPEP3 in AMI has been scarcely studied. Our analysis findings demonstrated an important role of XPNPEP3 in AMI development, offering encouragement for further research. To support our theory, molecular docking experiments showed that compound DB06909 could well interact with XPNPEP3 protein, and may be an inhibitor of XPNPEP3, suggesting DB06909 maybe an effective drug for AMI treatment.
In this work, we analyzed the mRNA-miRNA regulatory network in a public AMI database. The findings of this work could improve the current knowledge of AMI progression, and we have provided XPNPEP3-based therapeutic agent and diagnostic model. However, this study had some limitations including the relatively small sample size, and the lack of study on potential molecular mechanisms of the XPNPEP3 gene and functional experiments.
Conclusions
Through mRNA-miRNA regulatory network analysis, we established an XPNPEP3 gene diagnostic model and determined that DB06909 may be used to accurately diagnose and treat patients with AMI.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1203/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1203/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1203/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Samsky MD, Morrow DA, Proudfoot AG, et al. Cardiogenic Shock After Acute Myocardial Infarction: A Review. JAMA 2021;326:1840-50. [Crossref] [PubMed]
- Herrmann J. Peri-procedural myocardial injury: 2005 update. Eur Heart J 2005;26:2493-519. [Crossref] [PubMed]
- Nian M, Lee P, Khaper N, et al. Inflammatory cytokines and postmyocardial infarction remodeling. Circ Res 2004;94:1543-53. [Crossref] [PubMed]
- Fiedler J, Thum T. MicroRNAs in myocardial infarction. Arterioscler Thromb Vasc Biol 2013;33:201-5. [Crossref] [PubMed]
- Xiao J, Shen B, Li J, et al. Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int J Clin Exp Med 2014;7:136-41.
- Yang W, Shao J, Bai X, et al. Expression of Plasma microRNA-1/21/ 208a/499 in myocardial ischemic reperfusion injury. Cardiology 2015;130:237-41. [Crossref] [PubMed]
- Liu H, Yang N, Fei Z, et al. Analysis of plasma miR-208a and miR-370 expression levels for early diagnosis of coronary artery disease. Biomed Rep 2016;5:332-6. [Crossref] [PubMed]
- Laggerbauer B, Engelhardt S. MicroRNAs as therapeutic targets in cardiovascular disease. J Clin Invest 2022;132:e159179. [Crossref] [PubMed]
- Chen Y, Yang W, Wang GN, et al. Circulating microRNAs, novel biomarkers of acute myocardial infarction: a systemic review. World J Emerg Med 2012;3:257-60. [Crossref] [PubMed]
- Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499-506. [Crossref] [PubMed]
- Liu X, Fan Z, Zhao T, et al. Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: An independent study of Han population. Exp Gerontol 2015;72:230-8. [Crossref] [PubMed]
- Zhou J, He S, Wang B, et al. Construction and Bioinformatics Analysis of circRNA-miRNA-mRNA Network in Acute Myocardial Infarction. Front Genet 2022;13:854993. [Crossref] [PubMed]
- Wu J, Li C, Lei Z, et al. Comprehensive Analysis of circRNA-miRNA-mRNA Regulatory Network and Novel Potential Biomarkers in Acute Myocardial Infarction. Front Cardiovasc Med 2022;9:850991. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Jin X, Liu L, Wu J, et al. A multi-omics study delineates new molecular features and therapeutic targets for esophageal squamous cell carcinoma. Clin Transl Med 2021;11:e538. [Crossref] [PubMed]
- Kawada JI, Takeuchi S, Imai H, et al. Immune cell infiltration landscapes in pediatric acute myocarditis analyzed by CIBERSORT. J Cardiol 2021;77:174-8. [Crossref] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61. [Crossref] [PubMed]
- Das Mukherjee D, Kumar NM, Tantak MP, et al. NMK-BH2, a novel microtubule-depolymerising bis (indolyl)-hydrazide-hydrazone, induces apoptotic and autophagic cell death in cervical cancer cells by binding to tubulin at colchicine - site. Biochim Biophys Acta Mol Cell Res 2020;1867:118762. [Crossref] [PubMed]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235-42. [Crossref] [PubMed]
- Kerstjens A, De Winter H. LEADD: Lamarckian evolutionary algorithm for de novo drug design. J Cheminform 2022;14:3. [Crossref] [PubMed]
- Mooers BHM, Brown ME. Templates for writing PyMOL scripts. Protein Sci 2021;30:262-9. [Crossref] [PubMed]
- Rakhshani H, Dehghanian E, Rahati A. Enhanced GROMACS: toward a better numerical simulation framework. J Mol Model 2019;25:355. [Crossref] [PubMed]
- Shen W, Song Z, Xiao Z, et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022;3:e36.
- Shi LY, Han YS, Chen J, et al. Screening and identification of potential protein biomarkers for the early diagnosis of acute myocardial infarction. Ann Transl Med 2021;9:743. [Crossref] [PubMed]
- Huo H, Dai X, Li S, et al. Diagnostic accuracy of cardiac magnetic resonance tissue tracking technology for differentiating between acute and chronic myocardial infarction. Quant Imaging Med Surg 2021;11:3070-81. [Crossref] [PubMed]
- Feng L, Yang Z, Chen S, et al. Diagnostic value of myocardial stress detection based on feature tracking MRI in patients with acute myocardial infarction. J Thorac Dis 2022;14:3454-61. [Crossref] [PubMed]
- Peterson SM, Thompson JA, Ufkin ML, et al. Common features of microRNA target prediction tools. Front Genet 2014;5:23. [Crossref] [PubMed]
- Quiles JM, Narasimhan M, Shanmugam G, et al. Differential regulation of miRNA and mRNA expression in the myocardium of Nrf2 knockout mice. BMC Genomics 2017;18:509. [Crossref] [PubMed]
- Wu J, Cao J, Fan Y, et al. Comprehensive analysis of miRNA-mRNA regulatory network and potential drugs in chronic chagasic cardiomyopathy across human and mouse. BMC Med Genomics 2021;14:283. [Crossref] [PubMed]
- Wang M, Luo J, Wan L, et al. Screening genes associated with myocardial infarction and transverse aortic constriction using a combined analysis of miRNA and mRNA microarray. Gene 2015;571:245-8. [Crossref] [PubMed]
- Chen YH, Zhong LF, Hong X, et al. Integrated Analysis of circRNA-miRNA-mRNA ceRNA Network in Cardiac Hypertrophy. Front Genet 2022;13:781676. [Crossref] [PubMed]
- O'Toole JF, Liu Y, Davis EE, et al. Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. J Clin Invest 2010;120:791-802. [Crossref] [PubMed]
- Kumar R, Kotapalli V, Naz A, et al. XPNPEP3 is a novel transcriptional target of canonical Wnt/β-catenin signaling. Genes Chromosomes Cancer 2018;57:304-10. [Crossref] [PubMed]


